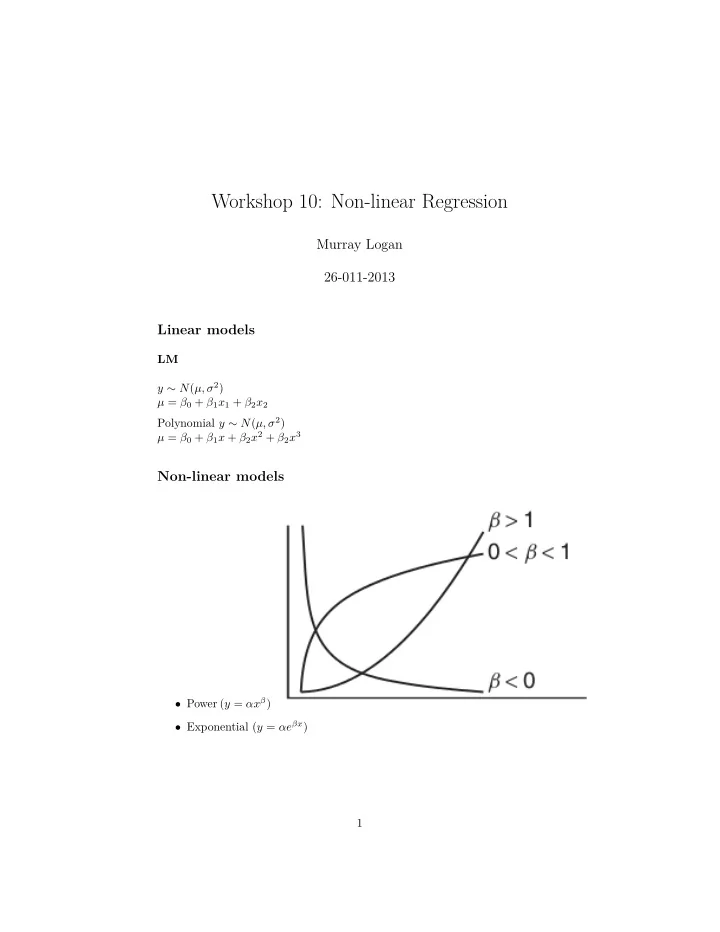

Workshop 10: Non-linear Regression Murray Logan 26-011-2013 Linear models LM y ∼ N ( µ, σ 2 ) µ = β 0 + β 1 x 1 + β 2 x 2 Polynomial y ∼ N ( µ, σ 2 ) µ = β 0 + β 1 x + β 2 x 2 + β 2 x 3 Non-linear models • Power ( y = αx β ) • Exponential ( y = αe βx ) 1
Non-linear models • Asymptotic exponential y = α +( β − α ) e − e γ x αx • Michaelis-Menton ( y = β + x ) 2
Non-linear models nls (y ~ a * exp (b * x), start = list (a = 1, b = 1)) Linear models LM y ∼ N ( µ, σ 2 ) µ = β 0 + β 1 x 1 + β 2 x 2 GLM y ∼ Pois ( µ, σ 2 ) g ( µ ) = β 0 + β 1 x 1 + β 2 x 2 Linear models • only permit parametric non-linearity – polynomials – nls Examples peake <- read.table ("../data/peake.csv", header = T, sep = ",", strip.white = T) head (peake) AREA SPECIES INDIV 1 516.0 3 18 2 469.1 7 60 3 462.2 6 57 4 938.6 8 100 5 1357.2 10 48 6 1773.7 9 118 ## @knitr Q4-a, fig.height=5.0, fig.width=5.0 library (car) scatterplot (SPECIES ~ AREA, data = peake) 3
Figure 1: plot of chunk unnamed-chunk-1 4
## @knitr Q4-2a peake.lm <- lm (SPECIES ~ AREA + I (AREA^2), data = peake) ## @knitr Q4-2b peake.nls <- nls (SPECIES ~ alpha * AREA^beta, data = peake, start = list (alpha = 0.1, beta = 1)) ## @knitr Q4-2c peake.nls1 <- nls (SPECIES ~ SSasymp (AREA, a, b, c), data = peake) ## @knitr Q4-3a peake.lmLin <- lm (SPECIES ~ AREA, data = peake) # calculate AIC for the linear model AIC (peake.lmLin) [1] 141.1 # calculate mean-square residual of the linear # model deviance (peake.lmLin)/ df.residual (peake.lmLin) [1] 14.13 AIC (peake.lm) [1] 129.5 # calculate mean-square residual of the power model deviance (peake.lm)/ df.residual (peake.lm) [1] 8.602 # assess the goodness of fit between linear and # polynomial models anova (peake.lmLin, peake.lm) 5
Analysis of Variance Table Model 1: SPECIES ~ AREA Model 2: SPECIES ~ AREA + I(AREAˆ2) Res.Df RSS Df Sum of Sq F Pr(>F) 1 23 325 2 22 189 1 136 15.8 0.00064 * — Signif. codes: 0 ‘ ’ 0.001 ’ ’ 0.01 ’ ’ 0.05 ’.’ 0.1 ’ ’ 1 # calculate AIC for the exponential asymptotic # model AIC (peake.nls) [1] 125.1 # calculate mean-square residual of the exponential # asymptotic model deviance (peake.nls)/ df.residual (peake.nls) [1] 7.469 # calculate AIC for the exponential asymptotic # model AIC (peake.nls1) [1] 125.8 # calculate mean-square residual of the exponential # asymptotic model deviance (peake.nls1)/ df.residual (peake.nls1) [1] 7.393 # assess the goodness of fit between polynomial and # power models anova (peake.nls, peake.nls1) Analysis of Variance Table Model 1: SPECIES ~ alpha * AREAˆbeta Model 2: SPECIES ~ SSasymp(AREA, a, b, c) Res.Df Res.Sum Sq Df Sum Sq F value Pr(>F) 1 23 172 2 22 163 1 9.14 1.24 0.28 6
opar <- par (mfrow = c (2, 2), mar = c (5, 5, 0, 0)) # first plot linear trend opar1 <- par (mar = c (5, 5, 0, 0)) plot (SPECIES ~ AREA, data = peake, ann = F, axes = F, type = "n") points (SPECIES ~ AREA, data = peake, col = "grey", pch = 16) xs <- with (peake, seq ( min (AREA), max (AREA), l = 1000)) ys <- predict (peake.lmLin, data.frame (AREA = xs), interval = "confidence") points (ys[, "fit"] ~ xs, col = "black", type = "l") points (ys[, "lwr"] ~ xs, col = "black", type = "l", lty = 2) points (ys[, "upr"] ~ xs, col = "black", type = "l", lty = 2) axis (1, cex.axis = 0.75) mtext ( expression ( paste ("Clump area", (dm^2))), 1, line = 3, cex = 1.2) axis (2, las = 1, cex.axis = 0.75) mtext ( expression ( paste ("Number of species (", phantom () %+-% 95, "%CI)")), 2, line = 3, cex = 1.2) box (bty = "l") par (opar1) # second plot polynomial trend opar1 <- par (mar = c (5, 5, 0, 0)) plot (SPECIES ~ AREA, data = peake, ann = F, axes = F, type = "n") points (SPECIES ~ AREA, data = peake, col = "grey", pch = 16) xs <- with (peake, seq ( min (AREA), max (AREA), l = 1000)) ys <- predict (peake.lm, data.frame (AREA = xs), interval = "confidence") points (ys[, "fit"] ~ xs, col = "black", type = "l") points (ys[, "lwr"] ~ xs, col = "black", type = "l", lty = 2) points (ys[, "upr"] ~ xs, col = "black", type = "l", lty = 2) axis (1, cex.axis = 0.75) mtext ( expression ( paste ("Clump area", (dm^2))), 1, line = 3, cex = 1.2) axis (2, las = 1, cex.axis = 0.75) mtext ( expression ( paste ("Number of species (", phantom () %+-% 95, "%CI)")), 2, line = 3, cex = 1.2) box (bty = "l") par (opar1) opar1 <- par (mar = c (5, 5, 0, 0)) 7
plot (SPECIES ~ AREA, data = peake, ann = F, axes = F, type = "n") points (SPECIES ~ AREA, data = peake, col = "grey", pch = 16) xs <- with (peake, seq ( min (AREA), max (AREA), l = 1000)) ys <- predict (peake.nls1, data.frame (AREA = xs)) se.fit <- sqrt ( apply ( attr (ys, "gradient"), 1, function(x) sum ( vcov (peake.nls1) * outer (x, x)))) points (ys ~ xs, col = "black", type = "l") points (ys + 2 * se.fit ~ xs, col = "black", type = "l", lty = 2) points (ys - 2 * se.fit ~ xs, col = "black", type = "l", lty = 2) axis (1, cex.axis = 0.75) mtext ( expression ( paste ("Clump area", (dm^2))), 1, line = 3, cex = 1.2) axis (2, las = 1, cex.axis = 0.75) mtext ( expression ( paste ("Number of species", ( phantom () %+-% 2 ~ SE), )), 2, line = 3, cex = 1.2) box (bty = "l") par (opar1) # we need to refit the model in order to get # gradient calculations from which se can be # calculated this requires that the gradient be # specified using the deriv3() function. grad <- deriv3 (~alpha * AREA^beta, c ("alpha", "beta"), function(alpha, beta, AREA) NULL) peake.nls <- nls (SPECIES ~ grad (alpha, beta, AREA), data = peake, start = list (alpha = 0.1, beta = 1)) ys <- predict (peake.nls, data.frame (AREA = xs)) se.fit <- sqrt ( apply ( attr (ys, "gradient"), 1, function(x) sum ( vcov (peake.nls) * outer (x, x)))) opar1 <- par (mar = c (5, 5, 0, 0)) plot (SPECIES ~ AREA, data = peake, ann = F, axes = F, type = "n") points (SPECIES ~ AREA, data = peake, col = "grey", pch = 16) points (ys ~ xs, col = "black", type = "l") # 2*SE equates approximately to 95% confidence # intervals technically it is qt(0.975,df)*SE # therefore this could also be labeled as 90%CI points (ys + 2 * se.fit ~ xs, col = "black", type = "l", lty = 2) points (ys - 2 * se.fit ~ xs, col = "black", type = "l", 8
lty = 2) axis (1, cex.axis = 0.75) mtext ( expression ( paste ("Clump area", (dm^2))), 1, line = 3, cex = 1.2) axis (2, las = 1, cex.axis = 0.75) mtext ( expression ( paste ("Number of species", ( phantom () %+-% 2 ~ SE), )), 2, line = 3, cex = 1.2) box (bty = "l") Figure 2: plot of chunk unnamed-chunk-1 Smoothers / splines • allow non-parametric non-linearity 9
• piecewise polynomial functions Piecewise regression Figure 3: Piecewise regression Smoothers / splines • allow non-parametric non-linearity • piecewise polynomial functions • joined by knots 10
Figure 4: 11
Piecewise regression Figure 5: Piecewise regression Piecewise regression y i = β 0 + β 1 ( x i ) + β 2 ( x i − cp ) y i = β 0 + β 1 I ( x i < x cp )( x i ) + β 2 I ( x i > x cp )( x i − x cp ) • single knot • determined by hypothesis 12
Figure 6: 13
Cubic spline Figure 7: Smoother • a function that relates Y to one or more X’s • types of smoothers – polynomials (parametric) – lowess, loess – splines ∗ cubic ∗ thin-plate 14
Smoother • non-parametric – no parameters – must be plotted to explore relationship • degree of smoothing Smoother - lowess Figure 8: 15
Degree of smoothing • λ • compromise – under and over smoothing – describe trend without focusing on outliers • model fit (residual sum of squares) • penalized for degree of smoothing (wiggliness) Degree of smoothing Generalized Cross-validation • create as many subsets as their are observations • fit all models with all λ • select best λ Generalized Additive Models LM y ∼ N ( µ, σ 2 ) µ = β 0 + β 1 x 1 + β 2 x 2 GLM y ∼ Pois ( µ, σ 2 ) g ( µ ) = β 0 + β 1 x 1 + β 2 x 2 Generalized Additive Models GLM y ∼ Pois ( µ, σ 2 ) g ( µ ) = β 0 + β 1 x 1 + β 2 x 2 GAM y ∼ Pois ( µ, σ 2 ) g ( µ ) = β 0 + f ( x 1 ) + f ( x 2 ) 16
Recommend
More recommend