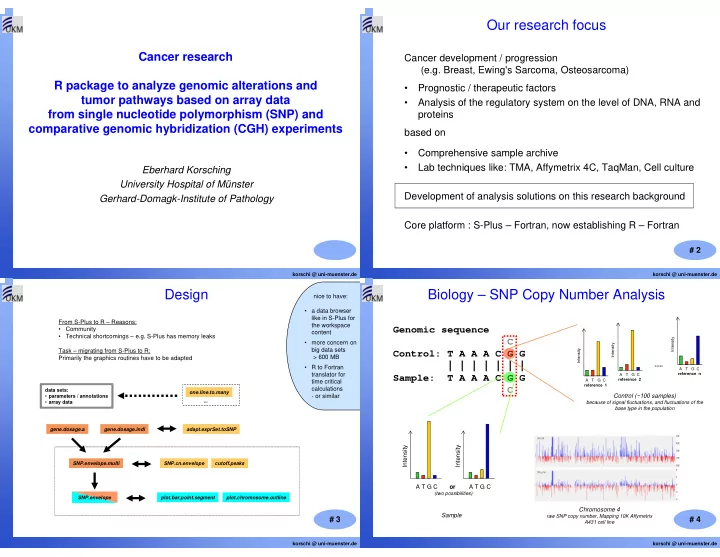

Our research focus Cancer research Cancer development / progression (e.g. Breast, Ewing's Sarcoma, Osteosarcoma) R package to analyze genomic alterations and • Prognostic / therapeutic factors tumor pathways based on array data • Analysis of the regulatory system on the level of DNA, RNA and from single nucleotide polymorphism (SNP) and proteins comparative genomic hybridization (CGH) experiments based on • Comprehensive sample archive • Lab techniques like: TMA, Affymetrix 4C, TaqMan, Cell culture Eberhard Korsching University Hospital of Münster Development of analysis solutions on this research background Gerhard-Domagk-Institute of Pathology Core platform : S-Plus – Fortran, now establishing R – Fortran # 2 korschi @ uni-muenster.de korschi @ uni-muenster.de Design Biology – SNP Copy Number Analysis nice to have: • a data browser like in S-Plus for From S-Plus to R – Reasons: the workspace Genomic sequence • Community content • Technical shortcomings – e.g. S-Plus has memory leaks C Intensity • more concern on Intensity big data sets Intensity Task – migrating from S-Plus to R: Control: T A A A C G G > 600 MB Primarily the graphics routines have to be adapted | | | | | | | ..... • R to Fortran A T G C translator for Sample: T A A A C G G reference n A T G C reference 2 A T G C time critical reference 1 C calculations data sets: one.line.to.many - or similar Control (~100 samples) • parameters / annotations ... • array data because of signal fluctuations, and fluctuations of the base type in the population gene.dosage.a gene.dosage.indi adapt.exprSet.toSNP Intensity Intensity SNP.envelope.multi SNP.cn.envelope cutoff.peaks A T G C or A T G C (two possibilities) SNP.envelope plot.bar.point.segment plot.chromosome.outline Chromosome 4 Sample raw SNP copy number, Mapping 10K Affymetrix # 3 # 4 A431 cell line korschi @ uni-muenster.de korschi @ uni-muenster.de
Visualization Take home message 1 1 1 1 - S-Plus to R is an easy task 1 1 1 1 93 Mb 1 51 Mb 1 1 1 48 Mb 124 Mb 61 Mb 92 Mb 45 Mb 35 Mb 59 Mb 40 Mb 51 Mb - SNPs are capable to replace the CGH technique 53 Mb - Old CGH data can be integrated 135 Mb 133 Mb 138 Mb 135 Mb 159 Mb 146 Mb 181 Mb 171 Mb 191 Mb 243 Mb 201 Mb 245 Mb - - - - - - - - - - - - Chr. 12 Chr. 10 Chr. 11 Chr. 8 Chr. 9 Chr. 6 Chr. 7 Chr. 5 SNPs: 644 SNPs: 545 Chr. 3 Chr. 4 SNPs: 544 SNPs: 610 Chr. 2 SNPs: 585 SNPs: 556 Chr. 1 SNPs: 793 SNPs: 816 SNPs: 780 SNPs: 813 SNPs: 962 SNPs: 881 Improvements in the Analysis Strategy Make Single Nucleotide Polymorphism Analysis a Powerful 1 Tool in the Detection and Characterization of Amplified Chromosomal Regions in Human Tumors 1 Eberhard Korsching a Konstantin Agelopolous a Hartmut Schmidt a Inka Buchroth a Georg Gosheger b Pia Wülfing c 1 59 Mb 1 16 Mb 16 Mb 1 Werner Boecker a Burkhard Brandt d Horst Buerger a 17 Mb 1 1 1 1 Institutes of a Pathology and b Orthopedics and c Department of Gynecology, University of Münster, Münster, and 16 Mb 22 Mb 38 Mb 12 Mb d Institute of Tumor Biology, University of Hamburg, Hamburg , Germany 28 Mb 27 Mb 12 Mb Pathobiology 2006;73: (DOI:10.1159/000093088) 155 Mb 76 Mb 65 Mb 62 Mb 47 Mb 50 Mb 100 Mb 89 Mb 81 Mb 114 Mb 106 Mb - - - - - - - - - - - Chr. 22 Chr. X Chr. 18 Chr. 19 Chr. 20 Chr. 21 Chr. 15 Chr. 16 Chr. 17 Chr. 14 Chr. 13 SNPs: 82 SNPs: 309 SNPs: 346 SNPs: 98 SNPs: 222 SNPs: 197 SNPs: 335 SNPs: 259 SNPs: 188 SNPs: 401 SNPs: 492 SNP copy number across genome Cooperation with: Walter Nadler MDA-MB-468 cell line, Mapping 10K Affymetrix, smoothing window: 40 The colored area indicates genetic alterations - Gains: green, losses: red Complex Systems Research Group, John von Neumann Institute for Computing, Research Centre Jülich, Germany & Computational Nano- and Biophysics Group, Department of Physics, Michigan Technological University, USA # 5 # 6 korschi @ uni-muenster.de korschi @ uni-muenster.de - Biology – CGH vs. SNP Analysis Comperative Genomic Hybridisation # 7 # 8 korschi @ uni-muenster.de korschi @ uni-muenster.de
Results I Results II 1 1 1 100% 80% 60% 40% 20% tumour DNA tumour DNA tumour DNA tumour DNA tumour DNA 51 Mb 51 Mb 51 Mb 1 1 1 191 Mb 191 Mb 1 191 Mb 1 16 Mb 16 Mb 16 Mb 16 Mb 16 Mb - 0 + - 0 + - 0 + 1 1 1 76 Mb 76 Mb 76 Mb 59 Mb 59 Mb 76 Mb 59 Mb 76 Mb - 0+ - 0+ - 0+ - 0+ - 0+ 159 Mb 159 Mb 159 Mb - 0 + - 0 + - 0 + Chr. 18 Chr. 18 Chr. 18 Chr. 18 Chr. 18 SNPs: 346 SNPs: 346 SNPs: 346 SNPs: 346 SNPs: 346 CGH SNP SNP SNP smoothing smoothing smoothing window 50 window 20 window 1 # 9 # 10 korschi @ uni-muenster.de korschi @ uni-muenster.de
Recommend
More recommend