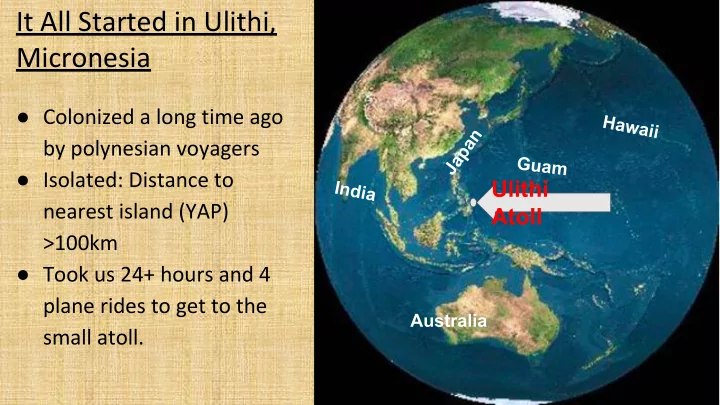

It All Started in Ulithi, Micronesia ● Colonized a long time ago Hawaii Japan by polynesian voyagers G u a m ● Isolated: Distance to Ulithi India nearest island (YAP) Atoll >100km ● Took us 24+ hours and 4 plane rides to get to the Australia small atoll.
Bio 80S: Applied Research Technologies: “Genetic Barcoding of Micronesian Fish” B Transition... To enable screen reader support, press Ctrl+Alt+Z To learn about keyboard shortcuts, press Ctrl+slash
Introduction We work on a research project of ● One People One Reef program, which is a collaboration between scientists and local communities in Micronesia for sustainable ocean management. The samples we used were fin ● clippings collected by local fisherman in Micronesia.
Class Goals Identify species being fished in Ulithi, and match to the names used ● locally Compare with samples of fish of same species from other countries ● Help identify populations or cryptic species ● Help focus future fisheries research ● Skill and knowledge building for the students ●
Introduction All lab work was done on ● Cabrillo College’s campus by students We fundraised for a new ● PCR machine for Cabrillo
What We Did DNA digestion/extraction ● PCR and Electrophoresis Gel ● Amplified DNA sequences, sent for sequencing at ● UC Berkeley Use computer programs to: ● Clean genetic sequences ○ ID species (Match genetic sequences to online ○ databases) Evaluate evolutionary relationships ○
Mitochondrial Gene CO1 Cytochrome Oxidase Subunit 1 ● Sometimes called COX 1 (in ● humans called MT-CO1) Gene region for DNA “barcoding” ● of almost all animal groups This means you can use this gene ● to tell species apart
Challenges Sample : Degradation ● Quality ○ Age ○ Made it hard to amplify the DNA ○ Tested by using gel to see quality of ○ DNA - showed some degradation We extracted DNA from ● Fish ○ Corals ○ Sea cucumbers ○
Challenges PCR troubleshooting : Nuanced differences in procedure ● for different species Unknown optimal annealing ● temperatures for each species
Challenges Human Error : Cross-contamination ● Working with several samples ○ at once Pipetting incorrect volumes ● Microscopic amounts ○
Results Extracted DNA of 93 samples ● Fish, sea cucumbers & corals ○ Amplified COI of 49 fish samples ● Found 14 fish species ● 10 were new to our genetic ● database! At the end of class, successfully ● amplified sea cucumber DNA The COI gene of Holothuria ○ atra To be continued next ○ semester!
Results: species added to genetic database (Fall ‘17): Siganus stellatus Bothus mancus Gymnosarda unicolor Brown spotted Flowery flounder Dogtooth tuna spinefoot Kyphosus vagiensis Brassy chub Stolephorus andhraensis (?) Anchovy species Siganus punctuatus Parupeneus barberinus Gold spotted spinefoot Dash-and-dot goatfish
Found in Fall ‘17, but also during Spring class Acanthurus lineatus Thunnus albacares Lined surgeonfish Yellowfin tuna Lethrinus rubrioperculatus Plectropomous laevis Spotcheek Emperor Blacksaddled coral grouper
Results: Samples collected in 2017 Katsuwonus pelamis Cheilopogon abei Benthosema fibulatum Skipjack tuna Abe’s flyingfish Lanternfish Juveniles!
Results (previous semester, Spring ‘17) Identified species: Acanthurus triostegus Caranx lugubris Acanthurus lineatus Caranx sexfasciatus Convict surgeonfish Black jack Lined surgeonfish Bigeye trevally Lethrinus rubrioperculatus Crenimugil crenilabis Kyphosus cinerascens Fringelip mullet Blue sea chub Spotcheek Emperor
Results (previous semester, Spring ‘17) Identified species (continued): Thunnus albacares Variola albimarginata Variola louti Plectropomus laevis White edged lyretail Yellowfin tuna Yellow edge lyretail Blacksaddled coral grouper Oxycheilinus unifasciatus Lutjanus kasmira Lutjanus gibbus Myripristis berndti Ring tail maori wrasse Blue striped snapper Humpback red snapper Blotch eye soldierfish
Results Phylogenetic tree of a Bacteria??? Stolephorus andhraensis (?)
Results Potential cryptic species Parupeneus barberinus Dash-and-dot goatfish
Graph: fish commonly caught in Ulithi We confirmed that the fish ● identified, are the ones reported, giving their database more credibility Mostly herbivores-this is found ● across all research angles (surveys, genetics, fisheries data) On top of that, some species we ● found appear to be either new species or cryptic
Applications ● Our genetic sampling can highlight unique genetic differences of the fish in the Micronesian region ● Our results are a starting point, that can help guide future research ● And this future research can inform sustainable management of fish populations
Experience Gained Hands on experience in a genetics ● laboratory Learn how to use academic software to ● analyze data Real world research experience ● Troubleshooting ○ Team building ○ DIPLOMA to certify these experiences!! ●
Thank you! Cabrillo College Biology Teachers: Eva Salas ● Nicole Crane ● John Carothers ● Yves Tan ● UCSC Professor Giacomo Bernardi
Questions
DNA extraction: DNEasy Blood and Tissue Kit Cell lysis: Digestion. Lysis & Incubate @ 56°C for 6hrs ● Purification: Prepares sample to bind to clay in spin ● column. Washing: Membrane w/ DNA, but has salts and ● proteins also Dry spin: Residual ethanol is BAD! ● Elution: Releases DNA into solution from spin ● column. We now have extracted DNA!!!
What’s so Special About CO1? The mitochondrial CO1 gene is a ● homologous gene found in all species. The mutation rate for the gene ● found in animals is high enough that species can be differentiated. Short enough to be easily ● sequenced (658 base pairs, depending on the species)
PCR and Gel Electrophoresis Using our extracted DNA samples we ran PCR ● Used specific primers to amplify COI gene for DNA barcoding ● PCR protocol with steps for denaturation, replication and annealing ● Run gel electrophoresis to visualize PCR products ● PCR products sent to UC Berkeley for sequencing ●
Computational Analysis: Geneious Sequence data analyzed in ● Geneious Align Forward and Reverse ● sequences Performed a multiple sequence ● alignment; cleaned and trimmed results to create consensus sequence
Results Phylogenetic tree of Thunnus albacares and closely related species
Computational Analysis: Genbank and BOLD Blast consensus sequences using genomic browsers (Genbank & BOLD) ● to determine what species we have by matching COI sequences Once species were identified, we downloaded sequences from Genbank ● and created neighbor- joining phylogenetic trees
Benefits Troubleshoot issues in lab ● procedure/results Research experience and ● expectations Team building in lab environment ● Practice with essential lab ● techniques
Recommend
More recommend