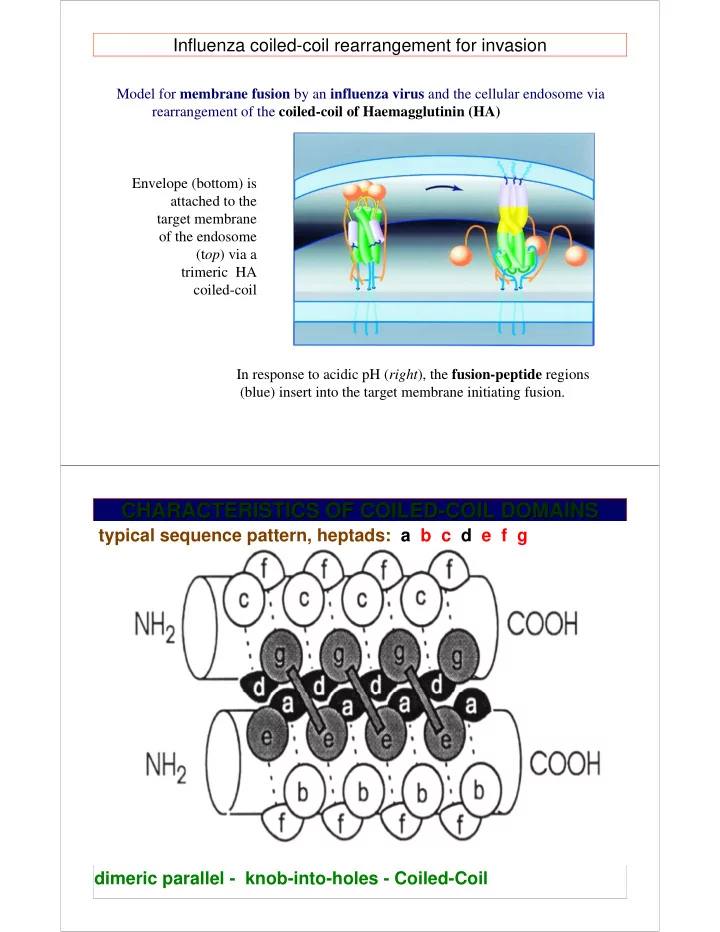

Influenza coiled-coil rearrangement for invasion Model for membrane fusion by an influenza virus and the cellular endosome via rearrangement of the coiled-coil of Haemagglutinin (HA) Envelope (bottom) is attached to the target membrane of the endosome (t op ) via a trimeric HA coiled-coil In response to acidic pH ( right ), the fusion-peptide regions (blue) insert into the target membrane initiating fusion. CHARACTERISTICS OF COILED- -COIL DOMAINS COIL DOMAINS CHARACTERISTICS OF COILED typical sequence pattern, heptads: a b c d e f g dimeric parallel - knob-into-holes - Coiled-Coil
COILED- -COIL CHARACTERISTICS COIL CHARACTERISTICS COILED • PERIODICAL ALTERNATION OF HYDROPHILIC AND HYDROPHOBIC AAs mainly characterised by the positions of the h'phobic AAs: • very variable length between ~ 11 and many hundreds of aas • frequently hphobic=>hphilic and vice-versa • WHAT IS THE BEST MODEL / METHOD FOR DESCRIBING AND RECOGNISING / PREDICTING THE OCCURRENCE OF THIS (HEPTAD) PATTERN ? COILED- -COIL DOMAINS COIL DOMAINS COILED dimeric Coiled-Coil with superhelical twist
COILED COILED- -COIL GROUPS COIL GROUPS • 1. TROPOMYOSINS • 2. MYOSINS • 3. INTERMEDIATE FILAMENTS • 4. DYNEINS • 6. LAMININS • 5. KINESINS • 7. SNARE PROTEINS • 8. LEU-ZIPPERS/TF • 9. OTHER PROTEINS CC CC- -PROBABILITY PROFILE AND PREDICTED DOMAINS PROBABILITY PROFILE AND PREDICTED DOMAINS predicted domain threshold Fusogenic Protein of simian Protein of simian parainfluenza parainfluenza Virus 5 Virus 5 Fusogenic y = P [ coiled − coil ( i )] = 1 − P [ state 0( i )]
DESIGNING A MODEL DESIGNING A MODEL • desired is a PARSING: • of sequences into coiled-coil and non coiled-coil (background) zones . - M - D - L - A - K - ..... DESIGNING A MODEL DESIGNING A MODEL Ca - Cb - Cc - Cd - Ce - Cf - Cg - Ca
DESIGNING A MODEL DESIGNING A MODEL Ca - Cb - Cc - Cd - Ce - Cf - Cg - Ca Cb - Cc - Cd - Ce - Cf - Cg - Ca - Cb DESIGNING A MODEL DESIGNING A MODEL the first simple model C1 C2 C7 C3 C6 C4 C5 arrows for the main alllowed transitions
DESIGNING A MODEL DESIGNING A MODEL C1 C2 C7 C3 C6 C4 C5 Design Issues: - LENGTH OF A COILED-COIL ZONE - FIRST AND LAST STRETCH ("CAPS") - REGULARITY OT THE HEPTAD PATTERN - COMPLEXITY AND PARAMETRISATION DESIGNING A MODEL DESIGNING A MODEL simplified structure of Marcoil
MARCOIL: architecture MARCOIL: architecture C-cap, 4 amino acids N-cap, 4 amino acids Internal Zone Marcoil Marcoil states: 10 groups, 9x7+1= 64 states states: 10 groups, 9x7+1= 64 states
PARAMETRISATION PARAMETRISATION 9*7+1=64 states 64*19 emission parameters ⇒ TYING reduces to 8*19=152 degrees of freedom for emission space (identical to COILS) 64*63 transitions parameters ? the important ones are much less, approx. 80 For the paper-version of Marcoil and a fairer comparison to Coils, the transition space was completely reparametrised and reduced to 3 degrees of freedom MARCOIL: MARCOIL: emiss. prob. . prob. emiss Example: state: a L: 25 .6% I : 1 3 .1% 56.9 % V: 11 .1% A: 7 .1% . K: 8 .5% . . W: 0 .2%
PARAMETERISATION PARAMETERISATION 3 parameters for all transition: i, t, r: i: initiation of a domain t: termination of a domain r: ratio btw "canonical" (heptad) transitions and "irregular" transitions In the productive mode: high i: frequent initiation of a domain high t: domain of shorter length In the decoding mode: The coiled-coil probability is increased by higher values of i. The ease with which a domain of length L is recognised is affected by i and t. For Viterbi decoding the dependency can be expressed quantitatively and the length-preference of the model is shifted towards shorter domains by increasing i and / or t. We believe (on theoretical and empirical grounds) that the same relationship holds qualitatively for Posterior Decoding. PARAMETERISATION PARAMETERISATION i ; t t 1 i
PARAMETERISATION PARAMETERISATION x i, t, r rx r = ratio between the minor and the major transitions (r << 1) HMM: decoding principle HMM: decoding principle WGP ARQLNES VKD INKM LER HP Sequence BBB CCCCCCC CCCCCCC CCC BB Labels Path1 000 abcdefg abcdefg abc 00 00c defgabc defgabc def g0 Path2 HMM-based model (“induced-fit principle”) a) VITERBI-decoding: of all possible states-path, we determine the best one (highest likelihood) (DP: log comlexity 10’000 => 8) b) POSTERIOR-decoding: at each single position, we determine the state with the highest probability (posterior to the given amino-acid sequence)
MARCOIL: decoding methods MARCOIL: decoding methods VITERBI POSTERIOR + uses all paths (weighted) - based only on the best path + SIMPLE + FAST - SLOWER / less STABLE + STABLE (log-space) + “CONSISTENT” PATH & MINIMAL LENGTH + EFFICIENT/NATURAL MEASURE OF STRINGENCY => VLR-VITERBI: slower CC CC- -PROBABILITY PROFILE AND PREDICTED DOMAINS PROBABILITY PROFILE AND PREDICTED DOMAINS predicted domain threshold Fusogenic Protein of simian Protein of simian parainfluenza parainfluenza Virus 5 Virus 5 Fusogenic y = P [ coiled − coil ( i )] = 1 − P [ state 0( i )]
LAB EXERCISE LAB EXERCISE We use a simplified model Starting with an initial set of parameters Train the parameters using a set of data by EM (Baum- Welch) Model File: Alphabet: ACDEFGHIKLMNPQRSTVWY ################################################ State BEGIN E: 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 T: BG 0.983 CCA 0.001 CCB 0.001 CCC 0.001 CCD 0.001 CCE 0.001 CCF 0.001 CCG 0.001 END 0.01 State BG E: 0.076 0.019 0.05 0.061 0.039 0.071 0.023 0.053 0.057 0.093 0.023 0.042 0.053 0.043 0.054 0.073 0.06 0.064 0.014 0.032 T: BG 0.983 CCA 0.001 CCB 0.001 CCC 0.001 CCD 0.001 CCE 0.001 CCF 0.001 CCG 0.001 END 0.01 State CCA E: 0.097 0.012 0.006 0.026 0.023 0.011 0.013 0.128 0.062 0.245 0.041 0.055 0.007 0.024 0.044 0.027 0.027 0.12 0.002 0.03 T: BG 0.001 CCA 0.001 CCB 0.983 CCC 0.001 CCD 0.001 CCE 0.001 CCF 0.001 CCG 0.001 END 0.01
Recommend
More recommend