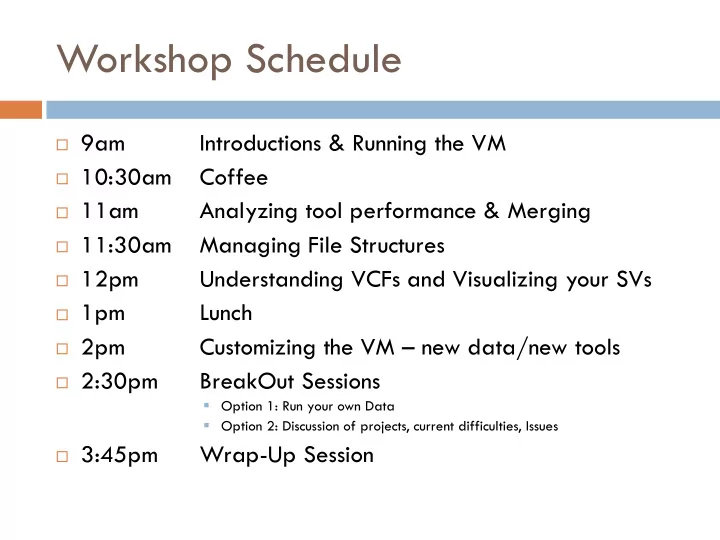

Workshop Schedule 9am Introductions & Running the VM 10:30am Coffee 11am Analyzing tool performance & Merging 11:30am Managing File Structures 12pm Understanding VCFs and Visualizing your SVs 1pm Lunch 2pm Customizing the VM – new data/new tools 2:30pm BreakOut Sessions Option 1: Run your own Data Option 2: Discussion of projects, current difficulties, Issues 3:45pm Wrap-Up Session
THE NEW FACE OF SV DETECTION Dr. Tiffanie Yael Maoz (Moss) Weizmann Institute of Science, Israel
Motivation for Work AllBio Research Consortium Project Test Case #2 – Identification of Large SVs Goal: Create a tool or pipeline which can identify large structural variants in multiple accessions of a genome
The Need for a Benchmark Many tools exist to predict structural variants (SVs), but have largely been developed and tested on human germline or somatic (e.g. cancer) variation. Benchmarking is needed to determine which tools are most suited non-human genomes.
SV Prediction Tools – Algorithm design
Procedure/Methodology Choose Non-Human Genome Choose SV tools to be tested Measure computational metrics Measure SV tool accuracy Optimize output for users Standardize process for future SV tool development
Non-Human Dataset Choice of Genome subject: Arabidopsis (Landsberg) Well annotated reference (TAIR v9/10) Validated Structural Variants (Landsberg – Ler ecotype) Diploid Homogenous Creation of semi-synthetic dataset Integration of INDEL calls into Reference genome 30x coverage More vs. Less ideal datasets
Tools Delly InGap (Java) BreakDancer GenomeSTRIP Tigra-SV GASV/GASVPro Hydra SVMerge SVDetect CNVSeq CNVnator Pindel PRISM
Popular SV tools
To do List Check quality of genomes and apply quality control as needed using fastqc Align reads with multiple alignment programs: BWA, Bowtie2, SOAP? Quality check the alignments Run SV programs on each alignment Format output as needed for visualization Test accuracy of output Write up the analysis
Day 1 Use Bam and Bowtie2 sorted bam files to run the following programs: Delly Breakdancer GASV-Pro SVDetect Pindel Prism Clever
Day 2 Completed the testing of the selected structural variation programs with bwa and bowtie2 alignments flagstat – Statistics for mapping quality for bwa and bowtie2 Used a script to compare the quality of the SV programs on the basis of precision, recall, novel calls, distance of the predicted from the true call and the difference in length. (Only examine SVs >10nt) Need add additional SV calls to our reference data set to be used in generating the synthetic reads Will use tool from the assemble-a-thon to create the synthetic reads in future (use same mean and standard deviation as was seen in the reference data) Working on generating a make file which should automate future runs on new test data and would properly format the VCF files for matrix analysis
Pipeline for Meta-Tool Virtual Machine (VM) Genomic Data Meta-Tool: Identification of the best tool for SV analysis SV analysis Data QC Reads alignment Alignments QC GASV Fastqc Samtools flagstat BWA-mem 0.7.5a Pindel Sickle Delly Prism Test dataset SVdetect Performance Optional: Merge Output format Clever Metrics Breakdancer Modified VCF Merged VCF Analysis PDF Downstream Options Genome Browser Select Tool(s) ESTs Develop/Refine Validate SVs Retro-elements X New Tools Small RNAs
Computational Performance SV’s Tool Threads Mem use on Mem use on CPU time CPU time Algorithm (reserved / Tair Human (mb) Tair (h:m.s) Human ran) (h:m.s) gasv 1 1058 594 0:02.08 0:01.20 Paired End IDVT delly 1 578 1236 0:15.02 0:03.18 Paired End & DVTP Split Read breakdancer 1 21.9 7 0:02.41 0:27.7 Paired End IDVT pindel 4 3500.5 5779 3:02.46 1:16.0 Split Read IDVP clever 1 - 4 238.7 1598 0:15.47 0:14.04 Paired End ID svdetect 1 - 2 172.3 3223 0:07.56 0:07.31 Paired End IDVTP SV types: Insertion (I), Deletion (D), Inversion (V), Translocation(T) , SNP (S), Duplication (P)
Measuring SV tool accuracy Measurements Precision & Recall Precision: percentage of correct calls among all calls Recall: percentage of true SV being called Across SV size classes 20-49nt 50-99nt 100-249nt 250-999nt 1000-50,000nt
Benchmarking Summary Tools vary in the calling ability across size classes Some tools are more adaptable to the Arabidopsis dataset Trade-off of many precise calls but often with more false positives There is benefit to using multiple tools, but the number of false positives would overwhelm the researcher with many non-true calls
Expectations for SV discovery Alkan, Coe & Eichler, 2011
Merging Package Merge Package Cluster and merge output Statistically deterimined clustering based on tool algorithms Calls reduced by as much as 70% VCF output format (suitable for IGV) Benefits of 7 callers with fewer false positives
Merge Package Overlay
AllBio Test Case #2 Hackers: W.Y. Leung, Sequencing Analysis Support Core, Leiden University Medical, The Netherlands Alexander Schoenhuth, Centrum Wiskunde & Informatica, Amsterdam, The Netherlands Tobias Marschall, Centrum Wiskunde & Informatica, Amsterdam, The Netherlands Leon Mei, Sequencing Analysis Support Core, Leiden University Medical, The Netherlands Yogesh Paudel, Animal breeding and genomics centre, Wageningen University, The Netherlands Laurent Falquet, University of Fribourg and Swiss Institute of Bioinformatics, Fribourg, Switzerland Tiffanie Yael Maoz (Moss), Weizmann Institute of Science, Rehovot, Israel Supported by: ALLBio Leadership with special thanks to Gert Vriend and Greg Rossier
Recommend
More recommend