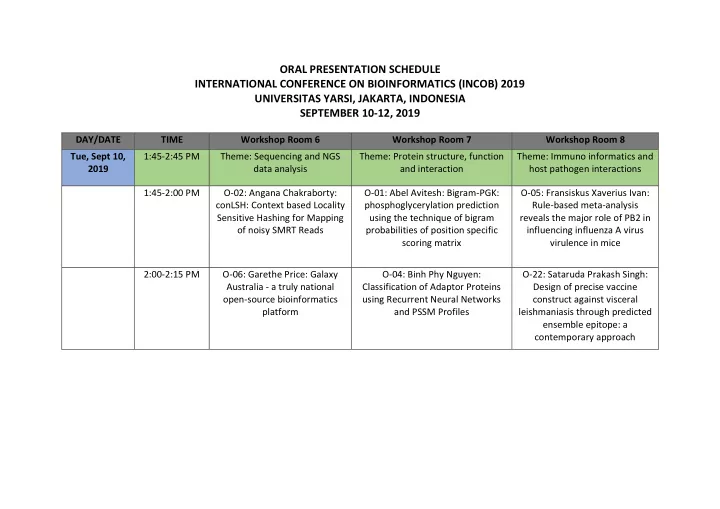

ORAL PRESENTATION SCHEDULE INTERNATIONAL CONFERENCE ON BIOINFORMATICS (INCOB) 2019 UNIVERSITAS YARSI, JAKARTA, INDONESIA SEPTEMBER 10-12, 2019 DAY/DATE TIME Workshop Room 6 Workshop Room 7 Workshop Room 8 Tue, Sept 10, 1:45-2:45 PM Theme: Sequencing and NGS Theme: Protein structure, function Theme: Immuno informatics and 2019 data analysis and interaction host pathogen interactions 1:45-2:00 PM O-02: Angana Chakraborty: O-01: Abel Avitesh: Bigram-PGK: O-05: Fransiskus Xaverius Ivan: conLSH: Context based Locality phosphoglycerylation prediction Rule-based meta-analysis Sensitive Hashing for Mapping using the technique of bigram reveals the major role of PB2 in of noisy SMRT Reads probabilities of position specific influencing influenza A virus scoring matrix virulence in mice 2:00-2:15 PM O-06: Garethe Price: Galaxy O-04: Binh Phy Nguyen: O-22: Sataruda Prakash Singh: Australia - a truly national Classification of Adaptor Proteins Design of precise vaccine open-source bioinformatics using Recurrent Neural Networks construct against visceral platform and PSSM Profiles leishmaniasis through predicted ensemble epitope: a contemporary approach
2:15-2:30 PM O-31: Yasubumi Sakakibara: An O-07: Hui Liu: MADOKA: An Ultra- O-44: Asif M Khan: Identification improved de novo genome fast Approach for Large-Scale of highly conserved, serotype- assembly of the common Protein Structure Similarity specific dengue virus sequences: marmoset genome yields Searching implications for vaccine design improved contiguity and increased mapping rates of sequence data 2:30-2:45 PM O-32: Younghi Lee: Differential O-45: Alhadi Bustamam: O-47: Shoba Ranganathan: alternative splicing regulation Performance of Rotation Forest Prediction of novel mouseTLR9 among hepatocellular Ensemble Classifier and Feature agonists using a random forest carcinoma with different risk Extractor in Predicting Protein approach factors Interactions Using Amino Acid Sequences 3:00-4:30 PM Theme: Genomics and Theme: Tools, databases and web NA Evolutionary Biology services in Bioinformatics 3:00-3:15 PM O-10: Jing Li: Genome-wide O-23: Sheng-Yao Su: EpiMOLAS: An identification, phylogeny, and Intuitive Web-based Framework for expression analysis of the SBP- Genome-wide DNA Methylation box gene family in Analysis Euphorbiaceae
3:15-3:30 PM O-48: Reeki Emirzal: O-28: Tsukasa Fukunaga: Logicome Phylogenetic analysis of Type IX Profiler: Exhaustive detection of Secretion System (T9SS) protein statistically significant logic components revealed that PorR relationships from comparative undergoes horizontal gene omics data transfer 3:30-3:45 PM O-41: Yen-Hua Huang: O-39: Xuan Zhang: JCDB: a Deconvolution of bulk gene comprehensive knowledge expression profiles from database for Jatropha curcas, an complex tissues to quantify emerging model for woody energy subsets of immune cells plants 3:45-4:00 PM O-18: Rajith Vidanaarachchi: O-46: Zhen Yang: CMTTdb: The IMPARO: Inferring Microbial Cancer Molecular Targeted Therapy Interactions through Parameter Database Optimisation 4:00-4:15 PM O35: Yung-Keun Kwon: Effects O-13: Kitty Lo: scDC: Single cell of ordered mutations on differential composition analysis dynamics in signaling networks
4:15-4:30 PM O-49: Jing Li: Genome-wide O-16: Pengyi Yang: scReClassify: identification, phylogeny, and post hoc cell type classification of expression analysis of the SBP- single-cell RNA-seq data box gene family in Euphorbiaceae Wed, Sept 11, 11:45-12:45 Theme: Mass spectrometry and Theme: Geneome wide association Theme: Machine learning, AI 2019 PM nano-bioinformatics studies (GWAS) and Biomarker and novel algorithms-I discovery 11:45-12.00 O-03: Anupam Nath Jha: O-14: Kyungsook Han: Finding O-11: Jinyan Li: Instance-Based PM Computational analysis of Silver prognostic gene pairs for cancer Learning for Personalized Cancer nanoparticle - human serum from patient-specific gene networks Diagnosis and Treatment albumin complex Planning 12:00-12.15 O-08: Jang-Jih Lu: Statistical O-25: Srinivasulu YS: O-12: Jinyan LI: DDI-PULearn: a PM Considerations and Machine Characterization of risk genes of novel positive-unlabeled Learning Approaches for Rapid autism spectrum disorders using learning method for large-scale Strain Typing of Staphylococcus gene expression profiles prediction of drug-drug haemolyticus based on Matrix- interactions Assisted Laser Desorption Ionization-Time-of Flight Mass Spectrometry
12:15-12:30 O-09: Jia-Ming Chang: O-34: Yun Zheng: The O-15: Le Ou-Yang: Predicting PM MS2CNN: Predicting MS/MS transcriptome variations of Panax Synthetic Lethal Interactions in spectrum based on protein notoginseng roots treated with Human Cancers using Graph sequence by Deep different forms of nitrogen Regularized Self-Representative Convolutional Neural Networks fertilizers Matrix Factorization 12:30-12:45 O-42: Tzong-Yi Lee: Rapid O-36: Zhixun Zhao: Identification of O-17: Pengyi Yang: PM Classification of Group B Lung Cancer Gene Markers through Autoencoder-based cluster Streptococcus Serotypes based kernel Maximum Mean Discrepancy ensembles for single-cell RNA- on Matrix-Assisted Laser and Information Entropy seq data analysis Desorption Ionization-Time of Flight Mass Spectrometry and Machine Learning Techniques Thu, Sept 12, 11:00-12:00 Theme: Network biology and Theme: Machine learning, AI and NA 2019 PM interaction networks novel algorithms-II 11:00-11:15 O-19: Rama Kalia: A module O-27: Susanto Rahardja: Enhancer AM refinement approach to find Identification and Classification functionally significant using One-Hot Encoding and communities in molecular Ensemble Convolutional Neural networks Networks
11:15-11:30 O-20: Rama Kalia: A module O-38: Dimitri Perrin: Improving AM refinement approach to find CRISPR guide design with consensus functionally significant approaches communities in molecular networks 11:30-11:45 O-30: Xioshi Zhong: GO2Vec: O-40: Ling Zou: Predicting AM Transforming GO Terms and synergistic drugs using gradient tree Proteins to Vector boosting based on features Representations Using Graph extracted from drug-protein Embeddings heterogeneous network 11:45-12:00 O-33: Young-Rae Cho: O-43: Kuo-Ching Liang: MetaVelvet- PM LePrimAlign: local entropy- DL: a MetaVelvet deep learning based alignment of PPI extension for de novo networks to predict conserved metagenomics assembly modules 1:00-1:45 PM Theme: Machine learning, AI Theme: Disease data modeling and NA and novel algorithms-III integrative Biology
1:00-1:15 PM O-21: Renmin Han: A novel O-24: Shobana Sundar: Rv0807, a constrained reconstruction putative phospholipase A2 of model towards high-resolution Mycobacterium tuberculosis; sub-tomogram averaging Elucidation through sequence analysis, homology modeling, molecular docking and molecular dynamics studies of potential substrates and inhibitors 1:15-1:30 PM O-37: Dhillon Sarinder Kaur: An O-26: Sudipto Saha: Computational Automated 3D Modelling approach to target USP28 for Pipeline for Constructing 3D regulating Myc Models of Monogenean Hardpart Using Machine Learning Techniques 1:30-1:45 PM O-50: Susanto Rahardja: O-29: Vivitri Dewi Prasastry: iEnhancer-ECNN: Identifying Structure-based Discovery of Novel enhancer and their strength Inhibitors of Mycobacterium using Ensemble of tuberculosis CYP121 from Convolutional Neural Networks Indonesian Natural Products
Recommend
More recommend