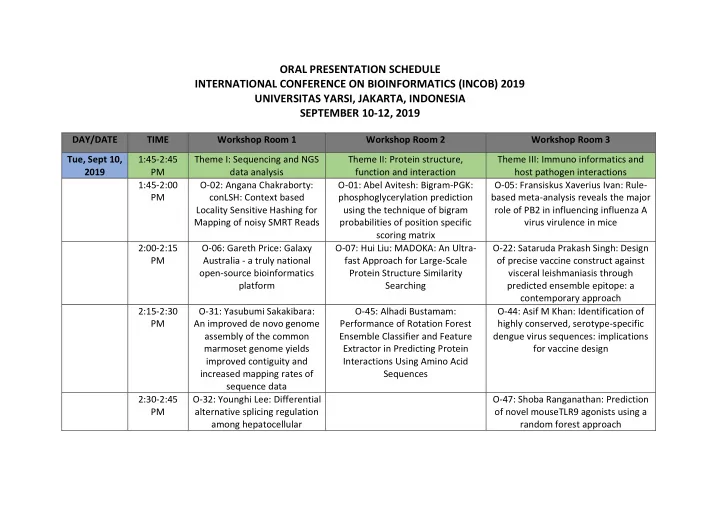

ORAL PRESENTATION SCHEDULE INTERNATIONAL CONFERENCE ON BIOINFORMATICS (INCOB) 2019 UNIVERSITAS YARSI, JAKARTA, INDONESIA SEPTEMBER 10-12, 2019 DAY/DATE TIME Workshop Room 1 Workshop Room 2 Workshop Room 3 Tue, Sept 10, 1:45-2:45 Theme I: Sequencing and NGS Theme II: Protein structure, Theme III: Immuno informatics and 2019 PM data analysis function and interaction host pathogen interactions 1:45-2:00 O-02: Angana Chakraborty: O-01: Abel Avitesh: Bigram-PGK: O-05: Fransiskus Xaverius Ivan: Rule- PM conLSH: Context based phosphoglycerylation prediction based meta-analysis reveals the major Locality Sensitive Hashing for using the technique of bigram role of PB2 in influencing influenza A Mapping of noisy SMRT Reads probabilities of position specific virus virulence in mice scoring matrix 2:00-2:15 O-06: Gareth Price: Galaxy O-07: Hui Liu: MADOKA: An Ultra- O-22: Sataruda Prakash Singh: Design PM Australia - a truly national fast Approach for Large-Scale of precise vaccine construct against open-source bioinformatics Protein Structure Similarity visceral leishmaniasis through platform Searching predicted ensemble epitope: a contemporary approach 2:15-2:30 O-31: Yasubumi Sakakibara: O-45: Alhadi Bustamam: O-44: Asif M Khan: Identification of PM An improved de novo genome Performance of Rotation Forest highly conserved, serotype-specific assembly of the common Ensemble Classifier and Feature dengue virus sequences: implications marmoset genome yields Extractor in Predicting Protein for vaccine design improved contiguity and Interactions Using Amino Acid increased mapping rates of Sequences sequence data 2:30-2:45 O-32: Younghi Lee: Differential O-47: Shoba Ranganathan: Prediction PM alternative splicing regulation of novel mouseTLR9 agonists using a among hepatocellular random forest approach
carcinoma with different risk factors 3:00-4:30 Theme IV: Genomics and Theme V: Tools, databases and web Theme VI: Network biology and PM Evolutionary Biology services in Bioinformatics interaction networks 3:00-3:15 O-10: Jing Li: Genome-wide O-23: Sheng-Yao Su: EpiMOLAS: An O-20: Rama Kalia: A module PM identification, phylogeny, and Intuitive Web-based Framework for refinement approach to find expression analysis of the SBP- Genome-wide DNA Methylation functionally significant communities box gene family in Analysis in molecular networks Euphorbiaceae 3:15-3:30 O-48: Reeki Emirzal: O-28: Tsukasa Fukunaga: Logicome O-30: Xiaoshi Zhong: GO2Vec: PM Phylogenetic analysis of Type Profiler: Exhaustive detection of Transforming GO Terms and Proteins IX Secretion System (T9SS) statistically significant logic to Vector Representations Using protein components revealed relationships from comparative Graph Embeddings that PorR undergoes omics data horizontal gene transfer 3:30-3:45 O-41: Yen-Jung Chiu: O-39: Xuan Zhang: JCDB: a O-33: Young-Rae Cho: LePrimAlign: PM Deconvolution of bulk gene comprehensive knowledge local entropy-based alignment of PPI expression profiles from database for Jatropha curcas, an networks to predict conserved complex tissues to quantify emerging model for woody energy modules subsets of immune cells plants 3:45-4:00 O-18: Rajith Vidanaarachchi: O-13: Yue Cao: scDC: Single cell O-04: Binh Phu Nguyen: Classification PM IMPARO: Inferring Microbial differential composition analysis of Adaptor Proteins using Recurrent Interactions through Neural Networks and PSSM Profiles Parameter Optimisation 4:00-4:15 O-35: Yung-Keun Kwon: O-16: Taiyun Kim: scReClassify: post O-27: Binh Phu Nguyen: Enhancer PM Effects of ordered mutations hoc cell type classification of single- Identification and Classification using on dynamics in signaling cell RNA-seq data One-Hot Encoding and Ensemble networks Convolutional Neural Networks
4:15-4:30 O-49: Sean Chun-Chang Chen: PM RNA editing-based classification of diffuse gliomas: predicting isocitrate dehydrogenase (IDH) mutation and chromosome 1p/19q codeletion Wed, Sept 11:45-12:45 Theme VII: Mass spectrometry Theme VIII: Genome wide Theme IX: Machine learning, AI and 11, 2019 PM and nano-bioinformatics association studies (GWAS) and novel algorithms-I Biomarker discovery 11:45-12.00 O-03: Zaved Hazarika: O-14: Kyungsook Han: Finding O-11: Md. Sarwar Kamal: Instance- PM Computational analysis of prognostic gene pairs for cancer Based Learning for Personalized Silver nanoparticle - human from patient-specific gene networks Cancer Diagnosis and Treatment serum albumin complex Planning 12:00-12.15 O-08: Jang-Jih Lu: Statistical O-25: Srinivasulu YS: O-12: Yi Zheng: DDI-PULearn: a novel PM Considerations and Machine Characterization of risk genes of positive-unlabeled learning method Learning Approaches for Rapid autism spectrum disorders using for large-scale prediction of drug-drug Strain Typing of gene expression profiles interactions Staphylococcus haemolyticus based on Matrix-Assisted Laser Desorption Ionization- Time-of Flight Mass Spectrometry 12:15-12:30 O-09: Yang Ming Lin: O-34: Yun Zheng: The O-15: Fan Lu: Predicting Synthetic PM MS2CNN: Predicting MS/MS transcriptome variations of Panax Lethal Interactions in Human Cancers spectrum based on protein notoginseng roots treated with using Graph Regularized Self- sequence by Deep different forms of nitrogen Representative Matrix Factorization fertilizers
Convolutional Neural Networks 12:30-12:45 O-42: Tzong-Yi Lee: Rapid O-36: Zhixun Zhao: Identification of O-17: Thomas Geddes: Autoencoder- PM Classification of Group B Lung Cancer Gene Markers through based cluster ensembles for single-cell Streptococcus Serotypes kernel Maximum Mean Discrepancy RNA-seq data analysis based on Matrix-Assisted and Information Entropy Laser Desorption Ionizat ion-Time of Flight Mass Spectrometry and Machine Learning Techniques Thu, Sept 12, 11:00-12:00 Theme X: Machine learning, AI NA NA 2019 PM and novel algorithms-II 11:00-11:15 O-38: Jacob Bradford: AM Improving CRISPR guide design with consensus approaches 11:15-11:30 O-40: Ling Zou: Predicting AM synergistic drugs using gradient tree boosting based on features extracted from drug-protein heterogeneous network 11:30-11:45 O-43: Kuo-Ching Liang: AM MetaVelvet-DL: a MetaVelvet deep learning extension for de novo metagenomics assembly
1:00-1:45 Theme XI: Machine learning, Theme XII: Disease data modeling NA PM AI and novel algorithms-III and integrative Biology 1:00-1:15 O-21: Lun Li: A novel O-24: Shobana Sundar: Rv0807, a PM constrained reconstruction putative phospholipase A2 of model towards high- Mycobacterium tuberculosis; resolution sub-tomogram Elucidation through sequence averaging analysis, homology modeling, molecular docking and molecular dynamics studies of potential substrates and inhibitors 1:15-1:30 O-37: Dhillon Sarinder Kaur: O-26: Sudipto Saha: Computational PM An Automated 3D Modelling approach to target USP28 for Pipeline for Constructing 3D regulating Myc Models of Monogenean Hardpart Using Machine Learning Techniques 1:30-1:45 O-29: Vivitri Dewi Prasastry: PM Structure-based Discovery of Novel Inhibitors of Mycobacterium tuberculosis CYP121 from Indonesian Natural Products 1:45-2:00 O-51: Asif M. Khan: A systematic PM bioinformatics approach for large- scale identification and characterization of host-pathogen shared sequences
Recommend
More recommend