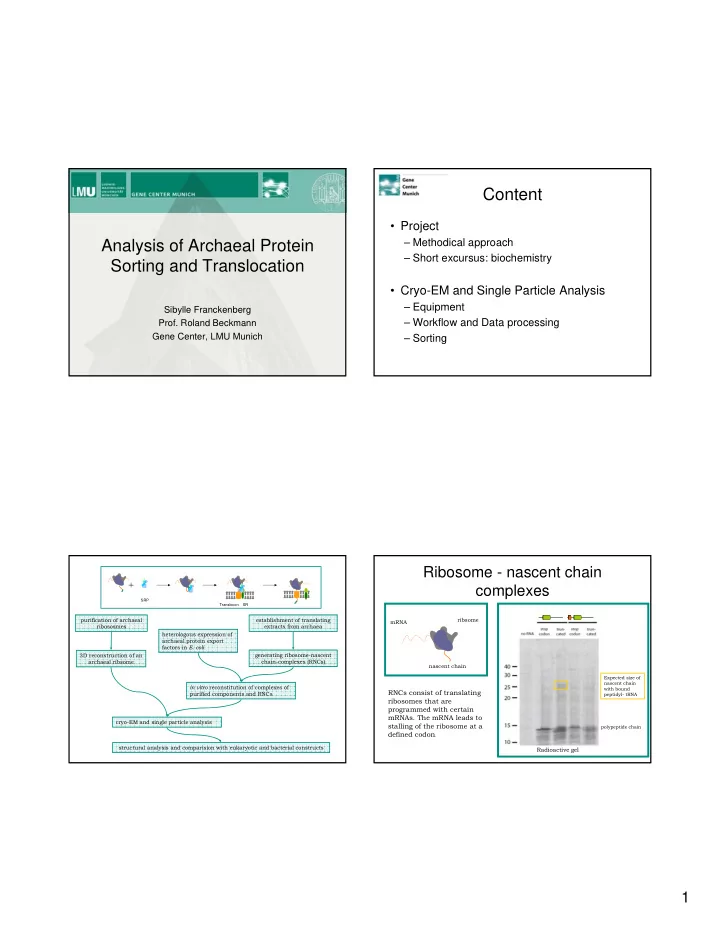
Content Project Analysis of Archaeal Protein Methodical approach - PDF document
Content Project Analysis of Archaeal Protein Methodical approach Short excursus: biochemistry Sorting and Translocation Cryo-EM and Single Particle Analysis Equipment Sibylle Franckenberg Workflow and Data processing
Content • Project Analysis of Archaeal Protein – Methodical approach – Short excursus: biochemistry Sorting and Translocation • Cryo-EM and Single Particle Analysis – Equipment Sibylle Franckenberg – Workflow and Data processing Prof. Roland Beckmann Gene Center, LMU Munich – Sorting Ribosome - nascent chain complexes SRP Translocon SR purification of archaeal establishment of translating ribsome mRNA ribososmes extracts from archaea heterologous expression of archaeal protein export factors in E. coli 3D reconstruction of an generating ribosome-nascent archaeal ribsome chain-complexes (RNCs) nascent chain Expected size of nascent chain in vitro reconstitution of complexes of with bound RNCs consist of translating purified components and RNCs peptidyl- tRNA ribosomes that are programmed with certain mRNAs. The mRNA leads to cryo-EM and single particle analysis stalling of the ribosome at a polypeptide chain defined codon . structural analysis and comparision with eukaryotic and bacterial constructs Radioactive gel 1
Cryo-sample preparation Archaeal protein export factors • Identification of factors based on BLAST analysis and the annotated genome from Thermococcus kodakarensis • Cloning into expression vectors • Heterologous expression in E. coli • Purification Quantifoil R 3/3 300 mesh Cu grids +2nm carbon on top • In vitro assembly and binding to RNCs Vitrobot Mark III, FEI Cryo-electron microscopes Digitalization of micrographs • Heidelberg Tango drum scanner Step size 4.3 µm � 1.2 Å/pixel Tecnai G2 Spirit Tecnai G2 Polara Titan Krios - 300 kV field emission - 80 - 300 kV tunable -120 kV transmission electron microscope gun field emission gun - computer-controlled - helium stage - cryo-autoloader sample cryostage - data collection on film stage - 2k x 2k CCD camera - 4k x 4k CCD-camera - 3.5 Å/pixel - Automated data acquisition 2
Workflow Pre-processing • CTF determination and visual • Sample preparation inspection of power spectra (Spider/Ctffind, Web) • Negative stain image: sample quality • Data collection on Tecnai Spirit G2 • Automated particle picking � low resolution reconstruction Negative stain (Signature) • Data collection on Tecnai Polara G2 � high resolution reconstruction • Manual particle selection (Web) • (Data collection on Titan Krios � high resolution reconstruction) Cryo Processing Processing • Resolution determination (Spider): • Alignment (Spider): Projection matching fourier shell correlation, cutoff 0.5 • Refinement (Spider): Projection matching • Initial back projection (Spider) 3
Dealing with inhomogenous Example I datasets Semi-supervised sorting – Two different volumes are used as initial reference for all particles – The dataset is segregated into two subpopulations depending on higher degree of similarity – The split datasets are individually used for back projection – Results are used as new references for the next round of sorting – The process continues until the particle number gets stable Different intensities of electron density of 50S and 30S subunits at lower contour level Example II 70S 50S Sorting 50S vs. 70S 60000 70S 50S 50000 40000 Particle number 30000 20000 10000 0 1 Sorting round 4
Validation of Sorting • Convergence of particle numbers Sorting programmed vs. idle 70S 60000 • Improving resolution (large datasets translating idle 50000 required) 40000 Particle number 30000 20000 • Projection with select files from one 10000 subpopulation and sorting-independent 0 angle files shows the same features like 1 Sorting round the sorted volume Outlook • Sample preparation: Initial reconstruction 20.5 Å, 20400 particles Pre-sorting reconstruction – RNCs/ribosomes and ligands 16 Å, 54500 50S subunit 12.4 Å, 10500 particles Low resolution data • Data interpretation: – Identification of conformational changes – tRNA positions (A/P/E-site) 70S ribosome – Ligand occupancy and 10.0 Å, 44000 particles conformational heterogeneity Idle ribosome Translating ribosome 14.0 Å, 20700 particles 11.5 Å, 23800 particles 5
Thank you • Beckmann lab – Dr. Thomas Becker • Wilson lab • Prof. Dr. Michael Thomm, University of Regensburg Early Cryo-EM 6
Recommend
More recommend
Explore More Topics
Stay informed with curated content and fresh updates.