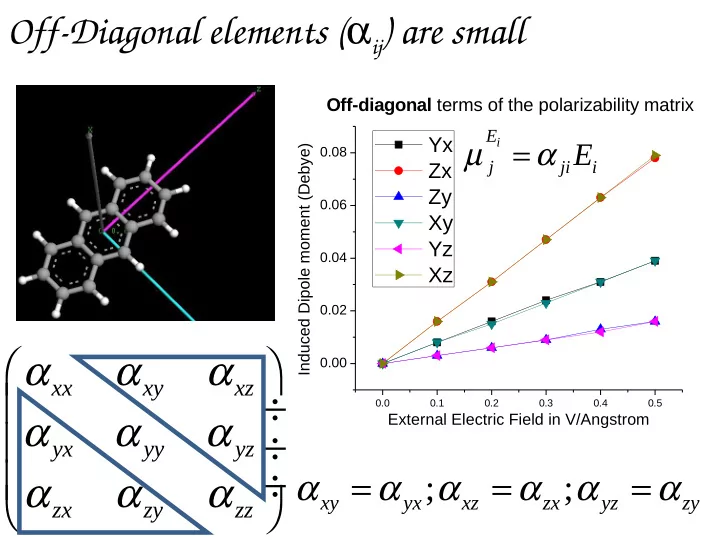

Off-Diagonal elements ( α ij ) are small Off-diagonal terms of the polarizability matrix u r E Yx µ = α i E Induced Dipole moment (Debye) 0.08 j ji i Zx Zy 0.06 Xy Yz 0.04 Xz 0.02 α α α 0.00 xx xy xz ÷ 0.0 0.1 0.2 0.3 0.4 0.5 α α α External Electric Field in V/Angstrom ÷ yx yy yz ÷ α = α α = α α = α α α α ; ; xy yx xz zx yz zy zx zy zz
Estimate relative magnitudes of polarizability components u u r u u r u u r u r = µ µ µ E E 0 = − = α E Ind u r u r µ E Average of the electronic α α α x ÷ ÷ Ix Polarizability ~ 1/3 xTrace of u r u r xx xy xz ÷ ÷ ÷ µ = α α α matrix – sum of diagonal elements. E ÷ y ÷ ÷ Iy yx yy yz (considering off-diagonal elements u r u r ÷ α α α ÷ ÷ relatively small) µ ÷ ÷ E zx zy zz z Iz u r u r u r u r = = E E 0 E E 0 µ − µ µ − µ i i i i α = α = j j i i ii ji E E i i u r u r u r u r = = E E 0 E E 0 µ = µ + α µ = µ + α i i i i E and E i i ii i j j ji i
Test case using Anthracene: Calculate µ ind by applying Ex, Ey, Ez u r = = E 0 µ i 0 Anthracene molecule aligned along z direction
The Hydrogen Bond The Hydrogen Bond
Boiling point of hydrides
Vibrational (IR) Spectra can measure the strength of a X-Y bond S-H O-H Frequency of oscillation of a bond is related to bond force constant – measure of bond strength
What is going on inside liquid water? Additional stabilization due to interaction between H atoms of one water molecule with the partially electronegative oxygen atom on another adjacent water molecule
Structure of liquid Water and Ice Commonality – H atom between two oxygen atoms; Distance between the H and two oxygen atoms not the same
Hydrogen bond involves…. Not all molecules can form hydrogen bond X-H…….Y-Z X-H…….Y-Z Molecule should have at least one hydrogen, bonded to a more electronegative atom. H-bond H-bond H-bond donor acceptor H-bond can form between two different molecules
Inter- and Intra-molecular H-Bonds Phenol-methanol Phenol-water Intra-molecular H-bonding
Hydrogen Bond Distances/Energies: Extremely large range! A D D H A D H H A O H O 1.0 1.8 2.8 1.0 1.9 2.9 O H N N H O 1.1 1.8 2.9 N H N 1.1 1.9 3.0 O H Cl 1.0 2.2 3.2 O H P 1.0 2.4 3.4
Concept and definition of H-bonding Under certain conditions an atom of hydrogen is attracted by rather strong forces to two atoms instead of only one, so that it may be considered to be acting as a bond between them. Linus Pauling (1939) IUPAC definition (2011) The hydrogen bond is an attractive interaction between a hydrogen atom from a molecule or a molecular fragment X–H in which X is more electronegative than H, and an atom or a group of atoms in the same or a different molecule, in which there is evidence of bond formation
Potential Energy for Hydrogen Bond complicated: poorly understood! Potential energy function for Hydrogen Bonding A B C D F G = − − − − − + V r ( ) 2 3 4 6 12 r r r r r r A - Electrostatics B - Ion-Dipole C - Dipole-Dipole D - Ion-Induced dipole F - Dipole-Induced dipole & Induced dipole -Induced dipole G - Repulsion Each hydrogen bond is unique Each hydrogen bond is unique
Importance of Intermolecular Forces and H-Bonds in Biology
3-DStructure of Proteins (Enzymes) held together: Intermolecular forces
Secondary structure of Proteins: H-Bonds play major role!
Held together by H-bonds!
The peptide alpha-Helix
The peptide beta-sheets
Nucleic acids – Double helical DNA
Structure of DNA
Base-pairing in nucleic acids •Each DNA base is hydrogen bonded to a base on the opposite strand forming a base pair • A bonds with T and G bonds with C forming complementary strands.
DNA Stabilization--H-bonding between DNA base pair stacks
Strength of Cellulose…..
Self-healing Rubber based solely on H-Bonds
Recommend
More recommend