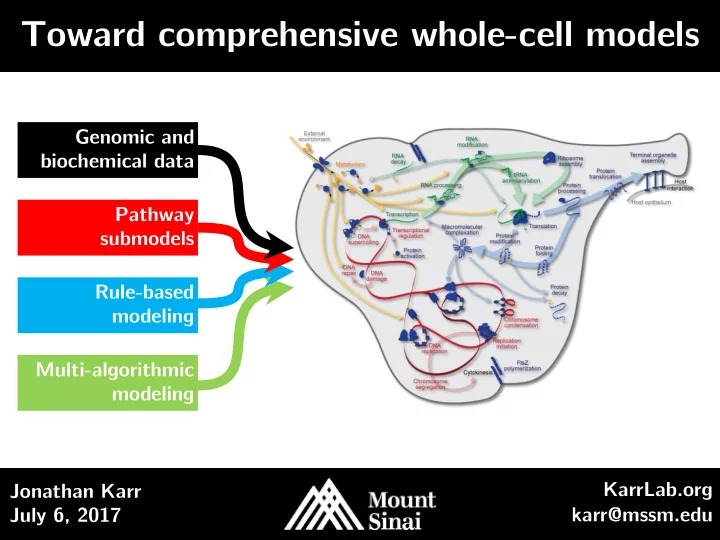

Toward comprehensive whole-cell models Genomic and biochemical data Pathway submodels Rule-based modeling Multi-algorithmic modeling KarrLab.org Jonathan Karr July 6, 2017 karr@mssm.edu
Outline Introduction to whole-cell (WC) modeling • What is a WC model? • Motivation • Challenges • Feasibility Methodology • Data aggregation • Data organization • Hybrid simulation New tools to accelerate WC modeling • Data aggregation • Model representation • Parallel simulation
Outline Introduction to whole-cell (WC) modeling • What is a WC model? • Motivation • Challenges • Feasibility Methodology • Data aggregation • Data organization • Hybrid simulation New tools to accelerate WC modeling • Data aggregation • Model representation • Parallel simulation
Features of whole-cell (WC) models Whole organism Dynamic Whole genome Stochastic including each gene Accurate Whole cell cycle Mechanistic Species-specific AGTC Karr et al., 2015
Outline Introduction to whole-cell (WC) modeling • What is a WC model? • Motivation • Challenges • Feasibility Methodology • Data aggregation • Data organization • Hybrid simulation New tools to accelerate WC modeling • Data aggregation • Model representation • Parallel simulation
Genome design requires WC models Biosensors Biofactories Tissue engineering
Example: drug biosynthesis
Example: drug biosynthesis
Example: drug biosynthesis
Example: drug biosynthesis
Example: drug biosynthesis
Example: drug biosynthesis
Example: drug biosynthesis
Personalized medicine requires WC models
Outline Introduction to whole-cell (WC) modeling • What is a WC model? • Motivation • Challenges • Feasibility Methodology • Data aggregation • Data organization • Hybrid simulation New tools to accelerate WC modeling • Data aggregation • Model representation • Parallel simulation
WC models are a grand challenge
Challenge: multiple time and length scales Growth Length Replication Transcription Metabolism Time
Challenge: heterogeneous data Protein expression Mass-spec, Western blot Transcription Single-cell variation RNA-seq Microscopy
Challenge: sparse data
Challenge: heterogeneous granularity Transcriptional regulatory Signaling Metabolic
Outline Introduction to whole-cell (WC) modeling • What is a WC model? • Motivation • Challenges • Feasibility Methodology • Data aggregation • Data organization • Hybrid simulation New tools to accelerate WC modeling • Data aggregation • Model representation • Parallel simulation
WC modeling is now feasible Genomic and biochemical data Pathway submodels Rule-based modeling Multi-algorithmic modeling
Extensive molecular data is available
Numerous predictors are available • miRNA targets: TargetScan • Operons: OperonPredictor • Protein half-lives: N-end rule • Protein localization: PSORT • Signal sequences: SignalP • Transcription start site: Promoter
Numerous databases are available
Model design tools are available MetaFlux
Model languages are available
Numerous pathway models are available
Numerous simulators are available Uptake FBA Composition Metabolism FBA Composition Transcription Stochastic binding Gene expression Translation Stochastic binding Gene expression Replication Chemical kinetics DNA sequence
Testing tools are available PRISM
Numerous other tools • Automated model construction • Model refinement • Parallel simulation • Calibration • Analysis and visualization • …
Outline Introduction to whole-cell (WC) modeling • What is a WC model? • Motivation • Challenges • Feasibility Methodology • Data aggregation • Data organization • Hybrid simulation New tools to accelerate WC modeling • Data aggregation • Model representation • Parallel simulation
Pathway modeling workflow 1. Choose a system to model 2. Determine the scope and granularity of the model 3. Determine the mathematical representation of the model 4. Reconstruct the species, reactions, rate laws, and rate parameters from the literature 5. Debug and calibrate the model by comparison to data 6. Test the model by comparison to independent data
Predictive modeling methodologies Boolean WC model Bolouri, 2000’s FBA Scope Palsson, 1990’s ODE Shuler, 1970’s PDE Gillespie Luthey-Schulten, 2011 Detail
Scaling pathway modeling to whole-cells • Aggregate more data – Accelerate data aggregation through automation – Organize input data using pathway/genome databases • Build models collaboratively using web-based tools – Define the semantic meaning of every model component – Track every assumption and data source • Describe models clearly – Explicitly describe the data used to build models – Describe models in terms of rules • Describe and simulate hybrid models
Scaling pathway modeling to whole-cells Genomics, bioinformatics ↔ Mechanistic modeling Pathway/genome databases ↔ Model design tools Polymers, sequences ↔ Rule-based modeling Stochastic modeling ↔ Steady-state modeling (FBA) Numerical simulation ↔ Big data analytics Model design tools ↔ Collaboration tools
WC modeling workflow
Aggregate data Genome Epigenome Transcriptome DNA-seq Meth-seq RNA-seq Proteome Metabolome Mass-spectrometry Mass-spectrometry Fraser et al., 1995; Kühner et al., 2009; Lluch-Senar et al., 2013; Maier et al., 2013; Yus et al. 2012
Organize input data Karr et al., 2013
Design submodels 1. Update RNA polymerase states 2. Calculate promoter affinities Fur HcrA GntR LuxR Spx Free Promoter Active Bound glpF dnaJ dnaK gntR trxB polC Bound 3. Bind RNA polymerase 4. Elongate and terminate transcripts Sequence AUGAUCCGUCUCUAAUGUCUAC Transcript UTCAACGUGAGGUAAUAAAGUC UCCACGAUGCUACUGUAUC GCCUCAUACUGCGGAU UUACGUAUCAGUGAUCAGUACU
Design pathway submodels Uptake FBA Composition Metabolism FBA Composition Transcription Stochastic events Gene expression Translation Stochastic events Gene expression Replication Chemical kinetics DNA sequence
Combine submodels Mass, shape Uptake FBA Composition Metabolite, RNA, protein counts Metabolism FBA Transcript, polypeptide Composition sequences Transcription Stochastic events Gene expression DNA polymerization, proteins, modifications Translation Stochastic events Gene expression FtsZ ring Replication Chemical kinetics DNA sequence Mammalian host States Submodels
Concurrently integrate submodels Uptake Uptake Uptake Metabolism Metabolism Metabolism Cell states Cell states Cell states Transcription Transcription Transcription Translation Translation Translation Replication Replication Replication 1 s
Calibrate model 1.Estimate individual parameters 2.Generate reduced models of individual pathways and to calibrate individual pathways 3.Refine joint parameter values using full models
Verify model against known biology Matches training data Matches theory Cell mass, volume Mass conservation Biomass composition Central dogma RNA, protein expression, half-lives Cell theory Superhelicity Evolution Matches published data No obvious errors Metabolite concentrations Plot model predictions DNA-bound protein density Manually inspect data Gene essentiality Compare to known biology
Verify model against known biology Matches training data Matches theory Cell mass, volume Mass conservation Biomass composition Central dogma RNA, protein expression, half-lives Cell theory Superhelicity Evolution Matches published data No obvious errors Metabolite concentrations Plot model predictions DNA-bound protein density Manually inspect data Gene essentiality Compare to known biology
Verify model against known biology Matches training data Matches theory Cell mass, volume Mass conservation Biomass composition Central dogma RNA, protein expression, half-lives Cell theory Superhelicity Evolution Matches published data No obvious errors Metabolite concentrations Plot model predictions DNA-bound protein density Manually inspect data Gene essentiality Compare to known biology
Verify model against known biology Matches training data Matches theory Cell mass, volume Mass conservation Biomass composition Central dogma RNA, protein expression, half-lives Cell theory Superhelicity Evolution Matches published data No obvious errors Metabolite concentrations Plot model predictions DNA-bound protein density Manually inspect data Gene essentiality Compare to known biology
Recommend
More recommend