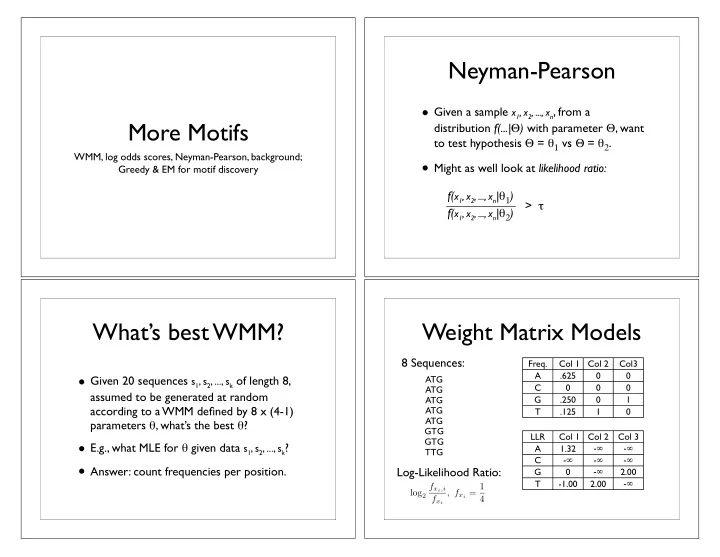

Neyman-Pearson • Given a sample x 1 , x 2 , ..., x n , from a More Motifs distribution f(...| � ) with parameter � , want to test hypothesis � = � 1 vs � = � 2 . WMM, log odds scores, Neyman-Pearson, background; • Might as well look at likelihood ratio: Greedy & EM for motif discovery f( x 1 , x 2 , ..., x n | � 1 ) f( x 1 , x 2 , ..., x n | � 2 ) > � What’s best WMM? Weight Matrix Models 8 Sequences: Freq. Col 1 Col 2 Col3 • Given 20 sequences s 1 , s 2 , ..., s k of length 8, A .625 0 0 ATG C 0 0 0 ATG assumed to be generated at random G .250 0 1 ATG according to a WMM defined by 8 x (4-1) ATG T .125 1 0 ATG parameters � , what’s the best � ? GTG LLR Col 1 Col 2 Col 3 GTG • E.g., what MLE for � given data s 1 , s 2 , ..., s k ? A 1.32 - � - � TTG - � - � - � C • Answer: count frequencies per position. Log-Likelihood Ratio: - � G 0 2.00 - � T -1.00 2.00 f x i ,i , f x i = 1 log 2 f x i 4
Non-uniform WMM: How “Informative”? Background Mean score of site vs bkg? • E. coli - DNA approximately 25% A, C, G, T • For any fixed length sequence x , let P(x) = Prob. of x according to WMM • M. jannaschi - 68% A-T, 32% G-C Q(x) = Prob. of x according to background LLR from previous • Recall Relative Entropy: LLR Col 1 Col 2 Col 3 example, assuming - � - � A .74 - � - � - � P ( x ) C � H ( P || Q ) = P ( x ) log 2 f A = f T = 3 / 8 - � G 1.00 3.00 Q ( x ) x ∈ Ω f C = f G = 1 / 8 - � T -1.58 1.42 -H(Q||P) H(P||Q) • H(P||Q) is expected log likelihood score of a e.g., G in col 3 is 8 x more likely via WMM sequence randomly chosen from WMM ; than background, so (log 2 ) score = 3 (bits). -H(Q||P) is expected score of Background WMM Example, cont. Freq. Col 1 Col 2 Col3 For WMM, you can show (based on the A .625 0 0 assumption of independence between C 0 0 0 columns), that : G .250 0 1 T .125 1 0 H ( P || Q ) = � i H ( P i || Q i ) Uniform Non-uniform where P i and Q i are the WMM/background LLR Col 1 Col 2 Col 3 LLR Col 1 Col 2 Col 3 A 1.32 - � - � A .74 - � - � distributions for column i. - � - � - � - � - � - � C C - � - � G 0 2.00 G 1.00 3.00 T -1.00 2.00 - � T -1.58 1.42 - � RelEnt .70 2.00 2.00 4.70 RelEnt .51 1.42 3.00 4.93
Pseudocounts How-to Questions • Given aligned motif instances, build model? • Are the - � ’s a problem? • Frequency counts (above, maybe with pseudocounts) • Certain that a given residue never occurs • Given a model, find (probable) instances? in a given position? Then - � just right • Scanning, as above • Else, it may be a small-sample artifact • Given unaligned strings thought to contain a • Typical fix: add a pseudocount to each motif, find it? (e.g., upstream regions for co- expressed genes from a microarray experiment) observed count—small constant (e.g., .5, 1) • Hard... next few lectures. • Sounds ad hoc ; there is a Bayesian justification Motif Discovery: Greedy Best-First Approach 3 example approaches [Hertz & Stormo] • Greedy search Input: usual “greedy” problems • Sequence s 1 , s 2 , ..., s k ; motif length I ; “breadth” d • Expectation Maximization Algorithm: • create singleton set with each length l • Gibbs sampler subsequence of each s 1 , s 2 , ..., s k • for each set, add each possible length l Note: finding a site of max relative entropy subsequence not already present in a set of unaligned sequences is NP-hard • compute relative entropy of each (Akutsu) • discard all but d best • repeat until all have k sequences
Expectation Maximization MEME Outline [MEME, Bailey & Elkan, 1995] Typical EM algorithm: Input (as above): • Given parameters � t at t th iteration, use • Sequence s 1 , s 2 , ..., s k ; motif length l ; background model; again assume one instance per sequence them to estimate where the motif instances (variants possible) are (the hidden variables) Algorithm: EM • Use those estimates to re-estimate the • Visible data: the sequences parameters � to maximize likelihood of • Hidden data: where’s the motif observed data, giving � t+1 � 1 if motif in sequence i begins at position j Y i,j = • Repeat 0 otherwise • Parameters � : The WMM Expectation Step Maximization Step (where are the motif instances?) (what is the motif?) E = 0 · P (0) + 1 · P (1) Find � maximizing expected value: � E ( Y i,j | s i , θ t ) = Y i,j Bayes Q ( θ | θ t ) = E Y ∼ θ t [log P ( s, Y | θ )] P ( Y i,j = 1 | s i , θ t ) = E Y ∼ θ t [log � k = i =1 P ( s i , Y i | θ )] P ( s i | Y i,j = 1 , θ t ) P ( Y i,j =1 | θ t ) E Y ∼ θ t [ � k = = i =1 log P ( s i , Y i | θ )] P ( s i | θ t ) E Y ∼ θ t [ � k � | s i | − l +1 = Y i,j log P ( s i , Y i,j = 1 | θ )] cP ( s i | Y i,j = 1 , θ t ) = i =1 j =1 � Y i,j E Y ∼ θ t [ � k � | s i | − l +1 = Y i,j log( P ( s i | Y i,j = 1 , θ ) P ( Y i,j = 1 | θ ))] c � � l i =1 j =1 k =1 P ( s i,j + k − 1 | θ t ) � k � | s i | − l +1 = = E Y ∼ θ t [ Y i,j ] log P ( s i | Y i,j = 1 , θ ) + C i =1 j =1 � k � | s i | − l +1 where c � is chosen so that � 1 3 5 7 9 11 ... � = Y i,j log P ( s i | Y i,j = 1 , θ ) + C j � i =1 j =1 Sequence i Y i,j = 1.
M-Step (cont.) Initialization � k � | s i | − l +1 � Q ( θ | θ t ) = Y i,j log P ( s i | Y i,j = 1 , θ ) + C i =1 j =1 1. Try every motif-length substring, and use as initial � a WMM with, say 80% of weight on Exercise: Show this is s 1 : ACGGATT. . . that sequence, rest uniform maximized by “counting” . . . GC. . . TCGGAC s k : letter frequencies over 2. Run a few iterations of each all possible motif � ACGG Y 1 , 1 � 3. Run best few to convergence instances, with counts CGGA Y 1 , 2 � Y 1 , 3 GGAT weighted by , again � Y i,j (Having a supercomputer helps) . . the “obvious” thing. . . . . � CGGA Y k,l − 1 � Y k,l GGAC
Recommend
More recommend