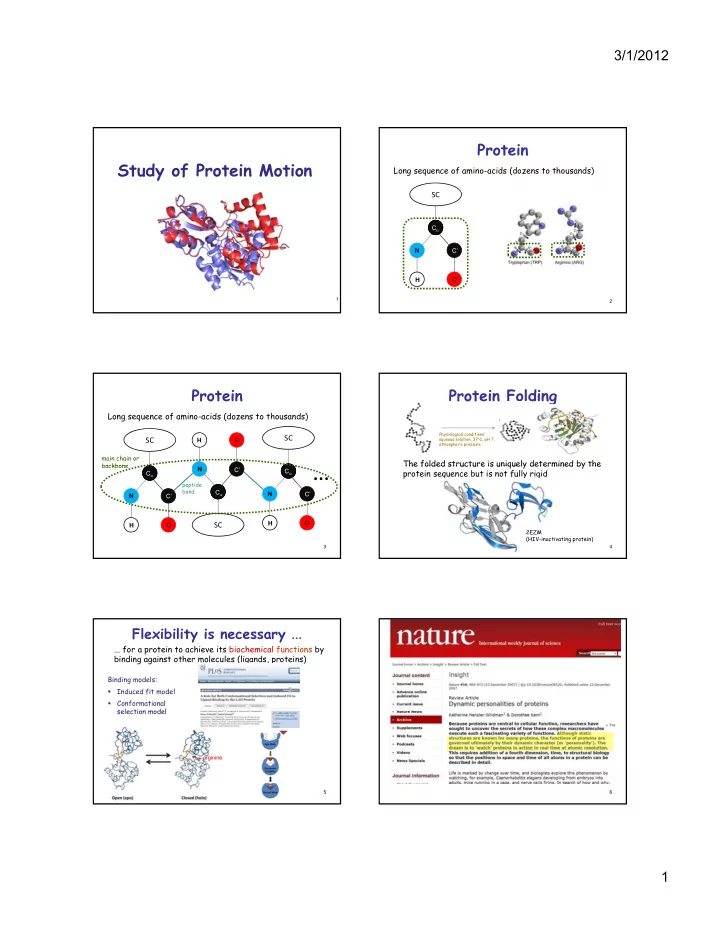

3/1/2012 Protein Study of Protein Motion Long sequence of amino-acids (dozens to thousands) SC C α C α C’ N O H 1 2 Protein Protein Folding Long sequence of amino-acids (dozens to thousands) Physiological conditions: SC O aqueous solution, 37 ° C, pH 7, SC H atmospheric pressure main chain or The folded structure is uniquely determined by the The folded structure is uniquely determined by the backbone backbone … N C’ C α C α protein sequence but is not fully rigid C α C α peptide C α C α bond C α C’ N C’ N O H O SC H 2EZM (HIV-inactivating protein) 3 4 Flexibility is necessary ... ... for a protein to achieve its biochemical functions by binding against other molecules (ligands, proteins) Binding models: Induced fit model � Conformational � selection model selection model arginine 5 6 1
3/1/2012 Experimental Methods Energy-Based Computer Simulation � Molecular Dynamics simulation Protein Data Bank (PDB): � Monte Carlo simulation Repository of folded structures of 74,732 � Others: proteins (August 2011) — coarse-grained force fields, — multi-scale modeling, � X-ray crystallography (65 195 entries) X ray crystallography (65,195 entries) — replica exchange, � high resolution, but one or few conformations — normal mode analysis, — elastic network models, … � NMR spectroscopy (9,014 entries) � Advantages: produce time-dependent information at � multiple conformations, but for small proteins atomic resolution � Drawbacks: huge running times, enormous amount of � Cryo-electron microscopy (373 entries) data, delicate setup � multiple conformations, but low resolution 7 8 Application of Robotics and Timescales of Protein Motion Motion Planning Techniques Bond/atomic Water Helix Fast correlated Slow 1. Kinematic/Geometric models of proteins vibration dynamics forms conf change conf change Develop algorithm-friendly models that directly encode dominant energy terms. 10 -15 10 -12 10 -9 10 -6 10 -3 10 0 femtosec picosec nanosec microsec millisec seconds MD step MD step one-day one day long long where we where we where we’d where we d 2. Kino-Geometric Conformational Sampling MD run MD run need to be love to be Use these models to sample conformations and [Pande] represent a protein’s folded state by a cloud of points. 3. Graph-Based Models of Protein Motion Transform a cloud representation into a probabilistic roadmap representing protein HIV-1 protease kinetics. 9 10 [courtesy L.E. Kavraki] Kinematic Model #1: How can one access directly to relevant timescales? Collection of independent atoms [Quick reminder: Kinematics studies the motion of objects without consideration Bond/atomic Water Helix Fast correlated Slow of the forces that cause the motion.] vibration dynamics forms conf change conf change Atoms can move independently, � i.e., all constraints between atoms 10 -15 10 -12 10 -9 10 -6 10 -3 10 0 are eventually represented in a force femtosec picosec nanosec microsec millisec seconds field function field function MD step MD step one day one-day long long where we where we where we’d where we d MD run MD run need to be love to be A conformation is defined by 3× n � [Pande] parameters (the coordinates of the � Simulating a protein over a nanosecond atom centers) timescale is like simulating human locomotion All motion frequencies can be � over a tiny fraction of a footstep, or like trying simulated to understand how to reach the Moon by jumping 1.5 feet in the air. 11 12 2
3/1/2012 Kinematic Model #1: Over picosecond timescales bond lengths Collection of independent atoms and angles average to constants Atoms can move independently, � R H i.e., all constraints between atoms are eventually represented in a force field function field function C α … … 114dg 123dg F = F bonded + F non-bonded 1.47Å 1.47Å 1.53Å 1.32Å C N 121dg 125dg stretching bending torsion 114dg 123dg O H ξ 13 14 Kinematic Model #2: Kinematic Model #2: Linkage of connected atoms Linkage of connected atoms Bonded atoms are connected by Bonded atoms are connected by � � links of fixed lengths links of fixed lengths The only degrees of freedom The only degrees of freedom � � are the dihedral angles around are the dihedral angles around the simple bonds the simple bonds 15 16 Kinematic Model #2: Kinematic Model #2: Linkage of connected atoms Linkage of connected atoms Bonded atoms are connected by Free vibrations of the atoms Bonded atoms are connected by Free vibrations of the atoms � � � � links of fixed lengths can no longer be generated links of fixed lengths can no longer be generated The only degrees of freedom The linkage model The only degrees of freedom The linkage model � � � � are the dihedral angles around are the dihedral angles around — encodes terms of the force field, — encodes terms of the force field, the simple bonds the simple bonds — filters out free atomic vibrations, — filters out free atomic vibrations, — retains long timescale motions — retains long timescale motions How can one encode other terms of the force field into the linkage model? 17 18 3
3/1/2012 Van der Waals Forces Hydrogen Bonds � F non-bonded = F van der Waals + F Coulomb � F non-bonded = F van der Waals + F Coulomb Van der Waals forces between two atoms result from electronegative induced polarization effect(formation of electric (often N or O) dipoles). They are weak, except at close range. electronegative 12-6 Lennard-Jones potential: H-bonds stabilize secondary structure elements and tertiary structure In a folded conformation, atoms are densely � packed against one another. Small perturbations can result into large repulsive vdW terms. Atoms are modeled as hard spheres with � radii ≈ α × vdWradii, where α = 0.7 to 0.8 + no two hard spheres are allowed to overlap 19 20 (volume exclusion constraint) Hydrogen Bonds Hydrogen Bonds � F non-bonded = F van der Waals + F Coulomb � F non-bonded = F van der Waals + F Coulomb electronegative electronegative (often N or O) (often N or O) electronegative electronegative H-bond H-bonds rigidify portions of the H-bonds rigidify portions of the protein and create closed protein and create closed cycles in linkage model cycles in linkage model Protein fragment 22 21 Advantages/Drawbacks of Inverse Kinematics Problem Linkage Model � Fewer DOFs, hence smaller dimensionality of How does a change in the position of an the conformational space atom affect the rest of the protein? � Many force terms are directly encoded in representation, hence the model can’t create p motion that would violate these terms � Most high-frequency motions are de facto filtered out But: � Generating kinematically valid conformations can be more difficult 23 24 4
3/1/2012 Analogy with Robotics Analogy with Robotics 25 26 Checking Volume-Exclusion Inverse Kinematics Methods Constraint: Grid Method Null-space motion How to deform a subset of a protein with more than 6 ~vdW diameter φ and ψ angles without breaking cycles? SVD method: — Cycle closure constraints � F ( q ) = 0 Cycle closure constraints ( q ) 0 — Differentiation � J F × dq = 0 q — SVD of J F � Basis of tangent space at q Subdivide 3-space into � cubic cells Compute cell that � contains each atom center Cartesian space Represent grid as hashtable of variable � dihedral angles 27 28 Beyond Simulation 1. Kinematic/Geometric models of proteins Develop algorithm-friendly models that directly encode dominant energy terms. 2. Kino-Geometric Conformational Sampling Use these models to sample conformations and represent a protein’s folded state by a cloud of points. 3. Graph-Based Models of Protein Motion Transform a cloud representation into a probabilstic representing protein kinetics. 29 30 5
Recommend
More recommend