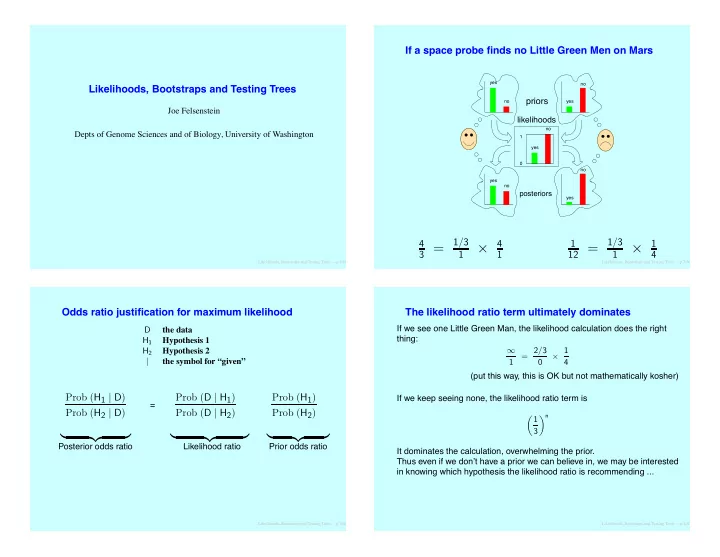

If a space probe finds no Little Green Men on Mars yes no Likelihoods, Bootstraps and Testing Trees priors no yes Joe Felsenstein likelihoods no Depts of Genome Sciences and of Biology, University of Washington 1 yes 0 no yes no posteriors yes 1 / 3 1 / 3 4 4 1 1 3 = 12 = × × 1 1 1 4 Likelihoods, Bootstraps and Testing Trees – p.1/60 Likelihoods, Bootstraps and Testing Trees – p.3/60 Odds ratio justification for maximum likelihood The likelihood ratio term ultimately dominates If we see one Little Green Man, the likelihood calculation does the right D the data thing: H 1 Hypothesis 1 Hypothesis 2 = 2 / 3 × 1 H 2 ∞ | the symbol for “given” 1 0 4 (put this way, this is OK but not mathematically kosher) Prob ( H 1 | D ) Prob ( D | H 1 ) Prob ( H 1 ) If we keep seeing none, the likelihood ratio term is = Prob ( H 2 | D ) Prob ( D | H 2 ) Prob ( H 2 ) � 1 � n 3 � �� � � �� � � �� � Posterior odds ratio Likelihood ratio Prior odds ratio It dominates the calculation, overwhelming the prior. Thus even if we don’t have a prior we can believe in, we may be interested in knowing which hypothesis the likelihood ratio is recommending ... Likelihoods, Bootstraps and Testing Trees – p.2/60 Likelihoods, Bootstraps and Testing Trees – p.4/60
Likelihood in Simple Coin-Tossing A likelihood curve Tossing a coin n times, with probability p of heads, the probability of A Likelihood curve in one parameter outcome HHTHTTTTHTTH is pp ( 1 − p ) p ( 1 − p )( 1 − p )( 1 − p )( 1 − p ) p ( 1 − p )( 1 − p ) p which is Ln (Likelihood) L = p 5 ( 1 − p ) 6 Plotting L against p to find its maximum: Likelihood length of a branch in the tree 0.0 0.2 0.4 0.6 0.8 1.0 p 0.454 Likelihoods, Bootstraps and Testing Trees – p.5/60 Likelihoods, Bootstraps and Testing Trees – p.7/60 Differentiating to find the maximum: Its maximum likelihood estimate Differentiating the expression for L with respect to p and equating the A Likelihood curve in one parameter derivative to 0, the value of p that is at the peak is found (not surprisingly) and the maximum likelihood estimate to be p = 5 / 11 : � 5 � ∂ L 6 p 5 ( 1 − p ) 6 = 0 ∂ p = p − 1 − p Ln (Likelihood) 5 − 11 p = 0 5 ˆ p = 11 length of a branch in the tree maximum likelihood estimate (MLE) Likelihoods, Bootstraps and Testing Trees – p.6/60 Likelihoods, Bootstraps and Testing Trees – p.8/60
The (approximate, asymptotic) confidence interval Contours of a likelihood surface in two dimensions A Likelihood curve in one parameter and the maximum likelihood estimate and confidence interval derived from it 1/2 the value of a chi − square length of branch 2 Ln (Likelihood) with 1 d.f. significant at 95% MLE 95% confidence interval length of a branch in the tree length of branch 1 maximum likelihood estimate (MLE) Likelihoods, Bootstraps and Testing Trees – p.9/60 Likelihoods, Bootstraps and Testing Trees – p.11/60 Contours of a likelihood surface in two dimensions Likelihood-based confidence set for two variables shaded area is the joint confidence interval length of branch 2 length of branch 2 height of this contour is less than at the peak by an amount equal to 1/2 the chi − square value with two degrees of freedom which is significant at 95% level length of branch 1 length of branch 1 Likelihoods, Bootstraps and Testing Trees – p.10/60 Likelihoods, Bootstraps and Testing Trees – p.12/60
Calculating the likelihood of a tree Likelihood-based confidence interval for one variable If we have molecular sequences on a tree, the likelihood is the product over sites of the data D [ i ] for each site (if those evolve independently): sites � Prob ( D [ i ] | T ) L = Prob ( D | T ) = i = 1 length of branch 2 With log-likelihoods, the product becomes a sum: sites � ln Prob ( D [ i ] | T ) ln L = ln Prob ( D | T ) = i = 1 height of this contour is less than at the peak by an amount equal to 1/2 the chi − square value with one degree of freedom which is significant at 95% level length of branch 1 Likelihoods, Bootstraps and Testing Trees – p.13/60 Likelihoods, Bootstraps and Testing Trees – p.15/60 Calculating the likelihood for site i on a tree Likelihood-based confidence interval for the other variable A C C C G t t 4 5 t1 t 2 t 3 y x length of branch 2 t 6 t7 ti are z "branch lengths", (rate X time) t w 8 Sum over all possible states (bases) at interior nodes: � � � � L ( i ) = Prob ( w ) Prob ( x | w , t 7 ) height of this contour is x y z w less than at the peak by an amount × Prob ( A | x , t 1 ) Prob ( C | x , t 2 ) Prob ( z | w , t 8 ) equal to 1/2 the chi − square value with one degree of freedom which is significant at 95% level × Prob ( C | z , t 3 ) Prob ( y | z , t 6 ) Prob ( C | y , t 4 ) Prob ( G | y , t 5 ) length of branch 1 Likelihoods, Bootstraps and Testing Trees – p.14/60 Likelihoods, Bootstraps and Testing Trees – p.16/60
Calculating the likelihood for site i on a tree and at the bottom of the tree: We use the conditional likelihoods: L ( i ) j ( s ) � L ( i ) π s L ( i ) = 0 ( s ) 0 These compute the probability of everything at site i at or above node j s on the tree, given that node j is in state s . Thus it assumes something ( s ) that we don’t know in practice – we compute these for all states s . (Felsenstein, 1973, 1981) At the tips we can define these quantities: if the observed state is (say) C , and having gotten the likelihoods for each site: the vector of L ’s is ( 0 , 1 , 0 , 0 ) sites � L ( i ) . L = If we observe an ambiguity, say R (purine), they are 0 i = 1 ( 1 , 0 , 1 , 0 ) Likelihoods, Bootstraps and Testing Trees – p.17/60 Likelihoods, Bootstraps and Testing Trees – p.19/60 The “pruning" algorithm: What does “tree space" (with branch lengths) look like? j k an example: three species with a clock trifurcation A B C not possible v v etc. j k t 1 t 1 l t 2 OK � � � t 2 L ( i ) Prob ( s j | s , v j ) L ( i ) � ( s ) = j ( s j ) when we consider all three possible topologies, the space looks like: s j � � � Prob ( s k | s , v k ) L ( i ) k ( s k ) × t1 t1 s k t2 t2 (Felsenstein, 1973; 1981). Likelihoods, Bootstraps and Testing Trees – p.18/60 Likelihoods, Bootstraps and Testing Trees – p.20/60
For one tree topology The graph of all trees of 5 species The space of trees varying all 2n − 3 branch lengths, each a nonegative number, defines an “orthant" (open corner) of a 2n − 3 -dimensional real space: C D D B A A B E E C B v 2 C E D C B B v D A A E wall 3 wall A C A v C B 8 D E v v 1 7 E B B C v 9 A A v 4 C D D E D v 6 B D E B F A A f v 9 l o o r D B C B v A C E D C A 5 C E D E E C A B C A B D A B E D C E D E A B D E C The Schoenberg graph (all 15 trees of size 5 connected by NNI’s) Likelihoods, Bootstraps and Testing Trees – p.21/60 Likelihoods, Bootstraps and Testing Trees – p.23/60 Through the looking-glass A data example: mitochondrial D-loop sequences Shrinking one of the n − 1 interior branches to 0, we arrive at a Bovine CCAAACCTGT CCCCACCATC TAACACCAAC CCACATATAC AAGCTAAACC AAAAATACCA Mouse CCAAAAAAAC ATCCAAACAC CAACCCCAGC CCTTACGCAA TAGCCATACA AAGAATATTA trifurcation: Gibbon CTATACCCAC CCAACTCGAC CTACACCAAT CCCCACATAG CACACAGACC AACAACCTCC Orang CCCCACCCGT CTACACCAGC CAACACCAAC CCCCACCTAC TATACCAACC AATAACCTCT B v 2 Gorilla CCCCATTTAT CCATAAAAAC CAACACCAAC CCCCATCTAA CACACAAACT AATGACCCCC v 3 v Chimp CCCCATCCAC CCATACAAAC CAACATTACC CTCCATCCAA TATACAAACT AACAACCTCC A C 8 v v 1 7 Human CCCCACTCAC CCATACAAAC CAACACCACT CTCCACCTAA TATACAAATT AATAACCTCC v 9 v 4 D v 6 F v TACTACTAAA AACTCAAATT AACTCTTTAA TCTTTATACA ACATTCCACC AACCTATCCA 5 TACAACCATA AATAAGACTA ATCTATTAAA ATAACCCATT ACGATACAAA ATCCCTTTCG E CACCTTCCAT ACCAAGCCCC GACTTTACCG CCAACGCACC TCATCAAAAC ATACCTACAA B v 2 CAACCCCTAA ACCAAACACT ATCCCCAAAA CCAACACACT CTACCAAAAT ACACCCCCAA v 3 v CACCCTCAAA GCCAAACACC AACCCTATAA TCAATACGCC TTATCAAAAC ACACCCCCAA A C 8 v v 4 v 1 7 B CACTCTTCAG ACCGAACACC AATCTCACAA CCAACACGCC CCGTCAAAAC ACCCCTTCAG B D v 2 v 2 CACCTTCAGA ACTGAACGCC AATCTCATAA CCAACACACC CCATCAAAGC ACCCCTCCAA v v 6 v v F 5 3 3 v C A v C 8 8 v v v 4 A v 1 5 7 v 9 E CACAAAAAAA CTCATATTTA TCTAAATACG AACTTCACAC AACCTTAACA CATAAACATA E D v v 1 v 9 7 v TCTAGATACA AACCACAACA CACAATTAAT ACACACCACA ATTACAATAC TAAACTCCCA 6 F v v 4 5 v 6 F CACAAACAAA TGCCCCCCCA CCCTCCTTCT TCAAGCCCAC TAGACCATCC TACCTTCCTA E D TTCACATCCG CACACCCCCA CCCCCCCTGC CCACGTCCAT CCCATCACCC TCTCCTCCCA CATAAACCCA CGCACCCCCA CCCCTTCCGC CCATGCTCAC CACATCATCT CTCCCCTTCA Here, as we pass “through the looking glass" we are also touch the space CACAAATTCA TACACCCCTA CCTTTCCTAC CCACGTTCAC CACATCATCC CCCCCTCTCA for two other tree topologies, and we could enter either. CACAAACCCG CACACCTCCA CCCCCCTCGT CTACGCTTAC CACGTCATCC CTCCCTCTCA CCCCAGCCCA ACACCCTTCC ACAAATCCTT AATATACGCA CCATAAATAA CA TCCCACCAAA TCACCCTCCA TCAAATCCAC AAATTACACA ACCATTAACC CA Likelihoods, Bootstraps and Testing Trees – p.22/60 Likelihoods, Bootstraps and Testing Trees – p.24/60 GCACGCCAAG CTCTCTACCA TCAAACGCAC AACTTACACA TACAGAACCA CA
Recommend
More recommend