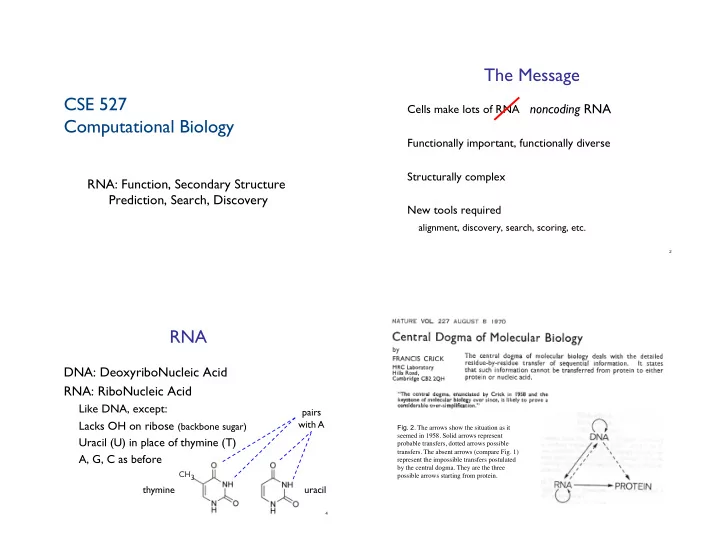

The Message " CSE 527 ! noncoding RNA " Cells make lots of RNA " Computational Biology " Functionally important, functionally diverse " Structurally complex " RNA: Function, Secondary Structure Prediction, Search, Discovery " New tools required " " alignment, discovery, search, scoring, etc. " 2 RNA " DNA: DeoxyriboNucleic Acid " RNA: RiboNucleic Acid " Like DNA, except: " pairs " Lacks OH on ribose (backbone sugar) " with A " Fig. 2 . The arrows show the situation as it seemed in 1958. Solid arrows represent Uracil (U) in place of thymine (T) " probable transfers, dotted arrows possible transfers. The absent arrows (compare Fig. 1) A, G, C as before " represent the impossible transfers postulated CH 3 " by the central dogma. They are the three possible arrows starting from protein. ! thymine " uracil " 4
Ribosomes " Ribosomes " 1974 Nobel prize to Romanian biologist George Palade (1912-2008) for discovery in mid 50’s " 50-80 proteins " 3-4 RNAs (half the mass) " Catalytic core is RNA " Atomic structure of the 50S Subunit from Of course, mRNAs and tRNAs Haloarcula marismortui . Proteins are shown in blue and the two RNA strands in orange (messenger & transfer RNAs) are ! and yellow. The small patch of green in the center of the subunit is the active site. � critical too " - Wikipedia � 7 " 8 " Watson, Gilman, Witkowski, & Zoller, 1992 “Classical” RNAs " Transfer RNA " The “adapter” coupling mRNA ! rRNA - ribosomal RNA (~4 kinds, 120-5k nt) " to protein synthesis. " tRNA - transfer RNA (~61 kinds, ~ 75 nt) " Discovered in the mid-1950s by ! RNaseP - tRNA processing (~300 nt) " Mahlon Hoagland (1921-2009, " snRNA - small nuclear RNA (splicing: U1, etc, 60-300nt) " left), Mary Stephenson, and Paul Zamecnik a handful of others " (1912-2009; Lasker award winner, right). !
Bacteria " RNA Secondary Structure: ! RNA makes helices too " Triumph of proteins " U � C � 80% of genome is coding DNA " A � A � C � G � Base pairs � Functionally diverse " G � C � A � " receptors " G � C � A � U � A � U � " motors " C � G � C � G � A � U � " catalysts " U � G � G � C � 5´ � " regulators (Monod & Jakob, Nobel prize 1965) " A � 3´ � A � A � A � C � U � " … " Usually single stranded " 13 " 11 Met Pathways " Proteins catalyze & regulate biochemistry " … " 14
Gene Regulation: The MET Repressor " Alberts, et al, 3e. The Riboswitch protein alternative way SAM " SAM " Grundy & Henkin, Mol. Microbiol 1998 Epshtein, et al., PNAS 2003 Winkler et al., Nat. Struct. Biol. 2003 DNA " Protein " Alberts, et al, 3e. " 17 " 16 Alberts, et al, 3e. Alberts, et al, 3e. The The Riboswitch Riboswitch protein protein alternatives alternatives way way SAM-II " SAM-III " SAM-I " SAM-I " SAM-II " Fuchs et al., NSMB 2006 Corbino et al., Genome Biol. 2005 Grundy, Epshtein, Winkler Grundy, Epshtein, Winkler Corbino et al., 18 " 19 " et al., 1998, 2003 et al., 1998, 2003 Genome Biol. 2005
Alberts, et al, 3e. The Riboswitch protein alternatives way SAM-III " SAM-I " SAM-II " SAM-IV " Grundy, Epshtein, Winkler Corbino et al., Fuchs et al., Weinberg et al., 20 " 21 " et al., 1998, 2003 Genome Biol. 2005 NSMB 2006 RNA 2008 Example: Glycine Regulation " How is glycine level regulated? " Plausible answer: " g gce protein g g g TF g DNA TF glycine cleavage enzyme gene transcription factors (proteins) bind to DNA to turn nearby genes on or off 22 " 23
6S mimics an ! The Glycine Riboswitch " open promoter " Actual answer (in many bacteria): ! Bacillus/ ! Clostridium " gce protein g g Actino- bacteria " g g 5 ! 3 ! gce mRNA E.coli DNA glycine cleavage enzyme gene Barrick et al. RNA 2005 Trotochaud et al. NSMB 2005 Mandal et al. Science 2004 Willkomm et al. NAR 2005 24 25 Vertebrates " Bigger, more complex genomes " <2% coding " boxed = Widespread, deeply conserved, structurally confirmed riboswitch sophisticated, functionally diverse, biologically But >5% conserved in sequence? " (+2 more) important uses for ncRNA throughout And 50-90% transcribed? " prokaryotic world. And structural conservation, if any, invisible (without proper alignments, etc.) " What’s going on? " 26 " Weinberg, et al. Nucl. Acids Res., July 2007 35: 4809-4819. �
Vertebrate ncRNAs " MicroRNA " mRNA, tRNA, rRNA, … of course " 1st discovered 1992 in C. elegans " 2nd discovered 2000, also C. elegans " PLUS: " and human, fly, everything between " 21-23 nucleotides " snRNA, spliceosome, snoRNA, teleomerase, literally fell off ends of gels " microRNA, RNAi, SECIS, IRE, piwi-RNA, Hundreds now known in human " XIST (X-inactivation), ribozymes, … " may regulate 1/3-1/2 of all genes " development, stem cells, cancer, infectious diseases,… " 29 " 30 siRNA " ncRNA Characteristics " “Short Interfering RNA” " Often low levels " Also discovered in C. elegans ! Can come from anywhere " Possibly an antiviral defense, shares Sense, antisense, introns, intergenic " machinery with miRNA pathways " Often poorly conserved " Allows artificial repression of most genes in CDS : neutral ~ 10 : 1 vs ncRNA : neutral ~ 1.2 : 1 " May suggest “transcriptional noise” " most higher organisms " Huge tool for biology & biotech " 31
Noise? " Conservation? " HOWEVER: " Neutral rate underestimated? " Sometimes capped, spliced, polyA+ " Promoters also evolving rapidly " Some known ncRNAs are intronic ! Sequence/function constraint for RNA ≠ CDS " (e.g. some miRNAs, all snoRNAs) " Alignments are suspect away from CDS " Sometimes very precisely localized ! Alignments are not optimized for RNA structure " to specific compartments, cell types, ! developmental stages, ! Despite all this, there is evidence for purifying (esp. dev & neuronal …) " selection on ncRNA promoters, splice sites, tissue- specific expression patterns, indels, … " Origin of Life? " Bottom line? " A significant number of “one-off” examples " Life needs " Extremely wise-spread ncRNA expression " " information carrier: DNA " At a minimum, a vast evolutionary substrate " " molecular machines, like enzymes: Protein " New technology (e.g. RNAseq) exposing " making proteins needs DNA + RNA + proteins " more " " making (duplicating) DNA needs proteins " How do you recognize an interesting one? " Horrible circularities! How could it have arisen in an abiotic environment? " Conserved secondary structure "
RNA replicase ! Origin of Life? " RNA can carry information, too " RNA double helix; RNA-directed RNA polymerase " RNA can form complex structures " RNA enzymes exist (ribozymes) " RNA can control, do logic (riboswitches) " The “RNA world” hypothesis: ! 1st life was RNA-based " 39 Johnston et al., Science, 2001 " Why is RNA hard to deal with? " The Glycine Riboswitch " G � A � A � A � A � A � A � A � G � A � U � C � G � U � U � C � U � C � G � A � C � C � U � C � G � G � U � A � C � G � G � U � G � C � A � A � G � G � G � G � A � G � C � A � U � C � G � C � C � G � G � Actual answer (in many bacteria): ! C � A � G � C � A � A � G � A � G � G � A � G � G � A � G � A � G � G � A � C � A � C � C � A � C � U � U � G � U � A � C � C � C � C � gce G � A � A � A � A � protein G � A � g G � C � U � G � g C � C � A � A � A � A � U � A � A � A � G � A � G � U � G � A � G � A � C � A � C � U � C � U � U � g G � U � U � G � U � C � G � U � C � U � C � U � G � G � C � g 5 ! 3 ! G � A � G � C � G � U � C � G � G � A � C � G � C � A � U � U � gce mRNA G � C � U � G � A � A � A � A � C � G � U � G � C � U � U � G � U � U � G � A � U � DNA glycine cleavage enzyme gene G � G � C � G � A: Structure often more important than sequence Mandal et al. Science 2004 50 51
Wanted " Task 1: ! Good structure prediction tools " Structure Prediction ! Good motif descriptions/models " Good, fast search tools " (“RNA BLAST”, etc.) " Good, fast motif discovery tools " (“RNA MEME”, etc.) " Importance of structure makes last 3 hard " 54 RNA Pairing " RNA Structure " Primary Structure: " Sequence " Watson-Crick Pairing " C - G " " ~ 3 kcal/mole " Secondary Structure: " Pairing " A - U " " ~ 2 kcal/mole " “Wobble Pair” G - U " " ~1 kcal/mole " Tertiary Structure: " 3D shape " Non-canonical Pairs (esp. if modified) " 56
Recommend
More recommend