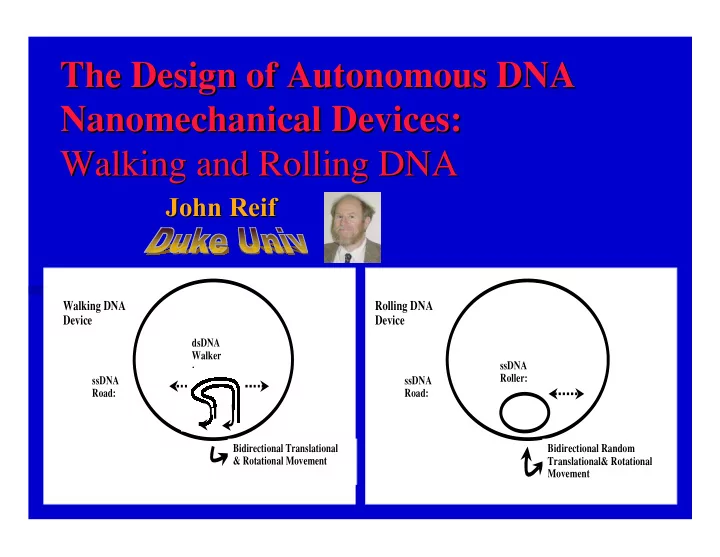

The Design of Autonomous DNA The Design of Autonomous DNA Nanomechanical Devices: Devices: Nanomechanical Walking and Rolling DNA Walking and Rolling DNA John Reif Reif John Walking DNA Rolling DNA Device Device dsDNA Walker ssDNA : Roller: ssDNA ssDNA Road: Road: Bidirectional Translational Bidirectional Random & Rotational Movement Translational& Rotational Movement
Hybridization of sticky single-strand DNA segments. Hybridization Ligation : If the sticky single-strand segments that anneal abut Ligation doubly stranded segments of DNA, you can use an enzymic ligation to concatenate these segments. reaction known as ligation
Prior Nanomechanical Devices built of DNA: · Seeman o used rotational transitions of dsDNA conformations between the B-form (right handed) to the Z-form (left-handed) controlled by ionic effector molecules and o extended this technique to be DNA sequence dependant Yurke and and Turberfield Turberfield · Yurke · o used a fuel DNA strands acting as a hybridization catalyst to generate a sequence of motions in another tweezers strand of DNA o extended this technique to be DNA sequence dependant o the two strands of DNA bind and unbind with the overhangs to alternately open and shut the tweezers. Other Related Work: Shapiro’s recent autonomous 2 state DNA computing machine · uses DNA ligase and two restriction enzyme • but distinct technical methods and goals (computation)
Bernard Yurke Yurke’ ’s s Molecular Tweezers (Bell Lab): Molecular Tweezers (Bell Lab): Bernard Composed of DNA and powered by DNA hybridization. Composed of DNA and powered by DNA hybridization. Two dsDNA dsDNA arms are connected by a arms are connected by a ssDNA ssDNA hinge hinge - Two - Two ssDNA ssDNA “ “handles handles” ” at the ends of the arms. at the ends of the arms. - Two - To close tweezers: To close tweezers: -Add a special “ “fuel fuel” ” strand of strand of ssDNA ssDNA. . -Add a special -The “ -The “fuel fuel” ” strand attaches to the handles and draws the two strand attaches to the handles and draws the two arms together. arms together.
B-Z DNA Nanomechanical Device [Seeman, 1999] D A B-Z Z-B A D
(a) (b) A B I II PX JX 2 PX JX 2 A B A B A B A B (b) (b) C D Z- B B- Z A B (a) (a) C D D C C D D C IV III D C PX PX PX (c) (d) (c) (d) JX 2 JX 2 JX 2 Nano-mechanical Rotational Transducers( -mechanical Rotational Transducers( Seeman Seeman, NYU) , NYU) Nano (a) DNA nanomechanical nanomechanical motor: Rotation via B-Z transition controlled by motor: Rotation via B-Z transition controlled by (a) DNA concentration of Co(NH 3 ) 6 C l 3 . concentration of Co(NH 3 ) 6 C l 3 . (b) Device switches between PX and JX 2 topological states of DNA controlled via (b) Device switches between PX and JX 2 topological states of DNA controlled via introduction of different strands, using Yurke YurkeÕ Õs s Molecular Tweezers. Molecular Tweezers. introduction of different strands, using (c) A test system where switching states alternates between a 'cis' 'cis' configuration configuration (c) A test system where switching states alternates between a (PX) and a 'trans' configuration (JX 2 (PX) and a 'trans' configuration (JX 2 ) . ) . (d) AFM pictures of four successive states through this system. (d) AFM pictures of four successive states through this system.
DNA Nanomechanical Nanomechanical Device ( Device (Hao Hao, Duke) , Duke) DNA 8 turns 180 _ _ 10.5 turns Walking Triangles: By binding the short red strand (top figure) versus the long red strand (bottom figure) the orientation of and distance between the triangular tiles is altered. These changes will be observable by AFM. Applications: Programmable state control for nanomechanical devices. Also as a visual output method.
NANOFACTO RY P- UP G- U P P- UP G- UP P- U P G- UP 1 1 1 (a) (a) 2 2 2 3 3 3 CYCLE 1 CYCLE 2 CYCLE 3 (b) (b) 1 1 1 2 2 2 3 3 3 Nanofactory device( device(Seeman Seeman, NYU): , NYU): Nanofactory PX/JX 2 devices with 3 cycles of configurations. PX/JX 2 devices with 3 cycles of configurations. (a) Nanogen Nanogen electrodes control release of hybridized strands into solution. electrodes control release of hybridized strands into solution. (a) (b) Three augmented device molecules mounted on an lattice. (b) Three augmented device molecules mounted on an lattice. Set strands of device labeled: P(urple urple)-up and G( )-up and G(reen reen)-up. )-up. Set strands of device labeled: P( Cycle 1: -Three G-up set strands on the device, Cycle 1: -Three G-up set strands on the device, -P-up set strands released into solution. -P-up set strands released into solution. Cycle 2: -G-up strands for molecules 1 and 3 released, Cycle 2: -G-up strands for molecules 1 and 3 released, -P-up strand for molecule 2 released. -P-up strand for molecule 2 released. Cycle 3: -P-up set strands for molecules 1 and 3 released, Cycle 3: -P-up set strands for molecules 1 and 3 released, -G-up set strand for molecule 2 released. -G-up set strand for molecule 2 released.
Patterned Immobilization of Environmentally-Responsive Patterned Immobilization of Environmentally-Responsive Peptides. Peptides. (on-going work in collaboration with (on-going work in collaboration with Chilkoti Chilkoti, Dept. of Biomedical Eng., Duke , Dept. of Biomedical Eng., Duke University.) University.) actuators that function in an aqueous environment . . Nanoscale actuators that function in an aqueous environment Nanoscale Molecular basis of nanoactuation nanoactuation: : Molecular basis of - ELPs ELPs are are Peptides that undergo a structural transition at a - Peptides that undergo a structural transition at a characteristic temperature. characteristic temperature. - The end-to-end distance of the ELP decreases by ~50% upon - The end-to-end distance of the ELP decreases by ~50% upon collapse of the ELP in response to its phase transition. collapse of the ELP in response to its phase transition. composed of : Hybrid materials composed of : Hybrid materials (a) self-assembling DNA nanostructures (a) self-assembling DNA nanostructures and and (b) elastin elastin-like peptides (ELP) -like peptides (ELP) (b) -Attachment of ELP to specific sites on DNA lattice results in arrays of peptide in -Attachment of ELP to specific sites on DNA lattice results in arrays of peptide in a monolayer monolayer of controlled density. of controlled density. a -May use layers of DNA sandwiched between layers of ELP. -May use layers of DNA sandwiched between layers of ELP.
Key restrictions on the use of prior DNA nanomechanical devices: Minor Restriction: They can only execute one type of motion (rotational or translational). Major Restriction: Prior DNA devices require environmental changes such as temperature cycling or bead treatment of biotin-streptavidin beads to make repeated motions. Our Technical Challenge: To make an autonomous DNA nanomechanical device · that executes cycles of motion (either rotational or translational or both) · without external environmental changes.
Our Results: Designs for the first autonomous DNA nanomechanical devices that execute cycles of motion without external environmental changes. Our two DNA Motor Devices: Walking DNA device, · O Uses ATP consumption by DNA ligase in conjunction with restriction enzyme operations. Rolling DNA device · O Uses hybridization energy These DNA devices translate across a circular strand of ssDNA and rotate simultaneously. Generate random bidirectional movements that acquire after n steps an expected translational deviation of O(n 1/2 ).
Energy sources that can fuel DNA movements: (i) ATP consumption by DNA ligase in conjunction with restriction enzyme operations (ii) DNA hybridization energy in trapped states (iii) kinetic (heat) energy
Walking DNA Autonomous Nanomechanical Device: requires no temperature changes. · Energetics: Uses ATP consumption by DNA ligase in conjunction with restriction enzyme operations. Achieves random bidirectional translational and rotational motion around a circular ssDNA strand. Walking DNA Device dsDNA Walker : ssDNA Road: Bidirectional Translational & Rotational Movement
Walking DNA Device Construction: The Road: A 0 A 1 A 2 …A n-1 A n … A...A ...A … · a circular repeating strand R of ssDNA · written in 5’ to 3’ direction from left to right. · consists of an even number n of subsequences, which we call steppingstones, indexed from 0 to n-1 modulo n. · The i th steppingstone consists of a length L (where L is between 15 to 20 base pairs) sequence A i of ssDNA. · In our constructions, the A i repeat with a period of 2. The i th Walker: W i : A i-1 A i A 0 A 1 … A i-1 A i A i+1 A unique a partial duplex DNA strand W i with 3’ ends i-1 and i that are hybridized to consecutive i-1 th and i th steppingstones A i-1 and A i ,
The Goal of the Device Construction: Bidirectional, translational movement both in the 5’ to 3’ direction (from left to right) and · vise versa (in the 3’ to 5’ direction) on the road. · The i th walker W i will reform to another partial duplex DNA strand called the i+1 th walker W i+1 which is: shifted one unit over to the left or the right. · Cycle back in 2 stages, so that W i+2 = W i for each stage i. · W i+1 : W i : A i A i+1 Step A i-1 A i A 0 A 1 … A i-1 A i A i+1 A 0 A 1 … A i-1 A i A i+1
Recommend
More recommend