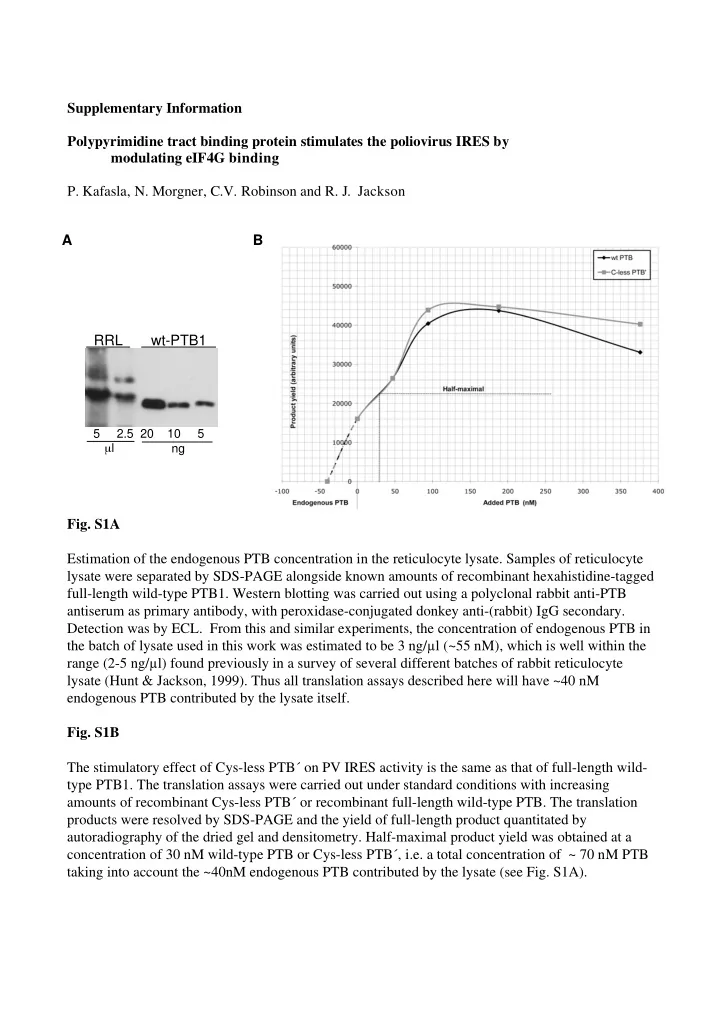

Supplementary Information Polypyrimidine tract binding protein stimulates the poliovirus IRES by modulating eIF4G binding P. Kafasla, N. Morgner, C.V. Robinson and R. J. Jackson A B RRL wt-PTB1 5 2.5 20 10 5 μ l ng Fig. S1A Estimation of the endogenous PTB concentration in the reticulocyte lysate. Samples of reticulocyte lysate were separated by SDS-PAGE alongside known amounts of recombinant hexahistidine-tagged full-length wild-type PTB1. Western blotting was carried out using a polyclonal rabbit anti-PTB antiserum as primary antibody, with peroxidase-conjugated donkey anti-(rabbit) IgG secondary. Detection was by ECL. From this and similar experiments, the concentration of endogenous PTB in the batch of lysate used in this work was estimated to be 3 ng/ � l (~55 nM), which is well within the range (2-5 ng/ � l) found previously in a survey of several different batches of rabbit reticulocyte lysate (Hunt & Jackson, 1999). Thus all translation assays described here will have ~40 nM endogenous PTB contributed by the lysate itself. Fig. S1B The stimulatory effect of Cys-less PTB´ on PV IRES activity is the same as that of full-length wild- type PTB1. The translation assays were carried out under standard conditions with increasing amounts of recombinant Cys-less PTB´ or recombinant full-length wild-type PTB. The translation products were resolved by SDS-PAGE and the yield of full-length product quantitated by autoradiography of the dried gel and densitometry. Half-maximal product yield was obtained at a concentration of 30 nM wild-type PTB or Cys-less PTB´, i.e. a total concentration of ~ 70 nM PTB taking into account the ~40nM endogenous PTB contributed by the lysate (see Fig. S1A).
Figure S2 A B C-less D242C D284C - C-less D242C D284C - : Fe(II)-PTB´ : Fe(II)-PTB´ - - - - - - - : p50 - - - - - - - : p50 + + + + + + T A G C T A G C - - + - - + - - + - - - + - - + - - + - : eIF4F : eIF4F 440- 530- 450- 460- 540- 550- 470- 560- 480- PVpr5 PVpr6 C D normalised band intensity normalised band intensity normalised band intensity normalised band intensity
Fig.S2 Native eIF4F complex and the eIF4G p50 central domain induce qualitatively identical changes in the cleavages in PV-IRES Domain V generated by Fe(II)-PTB´ derivatives. Tethered hydroxyl radical probing was carried out under standard conditions using 100 nM PV-IRES probe (nt 94-630), the designated Fe(II)-PTB´ derivatives at 350 nM, and 350 nM native reticulocyte eIF4F complex or 350 nM recombinant eIF4G p50 domain. Cleavage products were detected by primer extension using primers PVpr5 ( A ) and PVpr6 ( B ). (C, D) Quantitation of the designated cleavage product band intensities using ImageQuant TL software. The measured band intensities were normalised against the selected reference bands (arrowed in panels A and B) to adjust for loading variations, and the normalised band intensity is plotted. It is clear that the changes caused by the presence of native eIF4F complex are qualitatively the same, even though quantitatively greater, as those seen when eIF4G p50 is present. Note that the changes in cleavage band intensity caused by the presence of eIF4G p50 in this experiment were rather smaller than was typical (compare Figs. 5A and 5B). Part of the explanation is most likely due to the fact that the p50 concentration used in this experiment (350 nM) was half that tested in Figure 5, a change necessitated by the constraint that 350 nM was the maximum eIF4F concentration that was feasible. However, it is not clear that this concentration difference is the total explanation, and the possibility that this particular preparation of p50 was less active than the preparations used for Figure 5 cannot be ruled out. Consequently, no reliable conclusions can be drawn from the apparent quantitative differences between the changes induced by the presence by eIF4F and p50 in this figure. Supplementary References Hunt SL, Jackson RJ (1999) Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA 5 : 344-359.
Recommend
More recommend