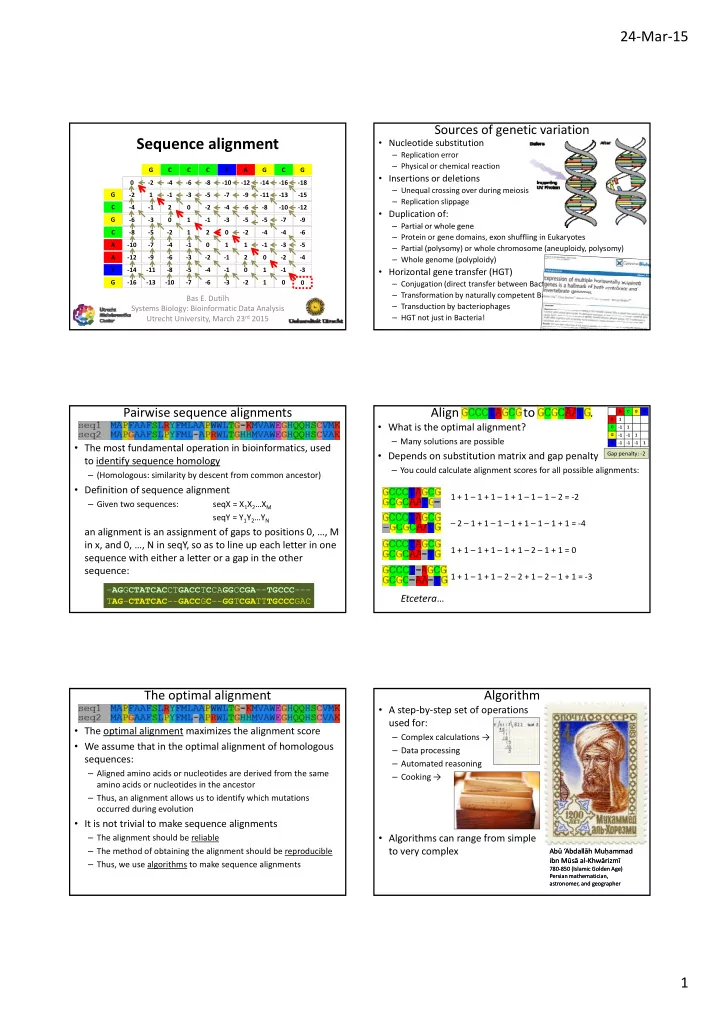

24 ‐ Mar ‐ 15 Sources of genetic variation Sequence alignment • Nucleotide substitution – Replication error – Physical or chemical reaction G C C C T A G C G • Insertions or deletions 0 0 ‐ 2 ‐ 2 ‐ 4 ‐ 4 ‐ 6 ‐ 6 ‐ 8 ‐ 8 ‐ 10 ‐ 10 ‐ 12 ‐ 12 ‐ 14 ‐ 14 ‐ 16 ‐ 16 ‐ 18 ‐ 18 – Unequal crossing over during meiosis G ‐ 2 ‐ 2 1 1 ‐ 1 ‐ 1 ‐ 3 ‐ 3 ‐ 5 ‐ 5 ‐ 7 ‐ 7 ‐ 9 ‐ 9 ‐ 11 ‐ 11 ‐ 13 ‐ 13 ‐ 15 ‐ 15 – Replication slippage C ‐ 4 ‐ 4 ‐ 1 ‐ 1 2 2 0 0 ‐ 2 ‐ 2 ‐ 4 ‐ 4 ‐ 6 ‐ 6 ‐ 8 ‐ 8 ‐ 10 ‐ 10 ‐ 12 ‐ 12 • Duplication of: G ‐ 6 ‐ 6 ‐ 3 ‐ 3 0 0 1 1 ‐ 1 ‐ 1 ‐ 3 ‐ 3 ‐ 5 ‐ 5 ‐ 5 ‐ 5 ‐ 7 ‐ 7 ‐ 9 ‐ 9 – Partial or whole gene – Partial or whole gene C ‐ 8 ‐ 8 ‐ 5 ‐ 5 ‐ 2 ‐ 2 1 1 2 2 0 0 ‐ 2 ‐ 2 ‐ 4 ‐ 4 ‐ 4 ‐ 4 ‐ 6 ‐ 6 – Protein or gene domains, exon shuffling in Eukaryotes A ‐ 10 ‐ 10 ‐ 7 ‐ 7 ‐ 4 ‐ 4 ‐ 1 ‐ 1 0 0 1 1 1 1 ‐ 1 ‐ 1 ‐ 3 ‐ 3 ‐ 5 ‐ 5 – Partial (polysomy) or whole chromosome (aneuploidy, polysomy) A ‐ 12 ‐ 12 ‐ 9 ‐ 9 ‐ 6 ‐ 6 ‐ 3 ‐ 3 ‐ 2 ‐ 2 ‐ 1 ‐ 1 2 2 0 0 ‐ 2 ‐ 2 ‐ 4 ‐ 4 – Whole genome (polyploidy) T ‐ 14 ‐ 14 ‐ 11 ‐ 11 ‐ 8 ‐ 8 ‐ 5 ‐ 5 ‐ 4 ‐ 4 ‐ 1 ‐ 1 0 0 1 1 ‐ 1 ‐ 1 ‐ 3 ‐ 3 • Horizontal gene transfer (HGT) G ‐ 16 ‐ 16 ‐ 13 ‐ 13 ‐ 10 ‐ 10 ‐ 7 ‐ 7 ‐ 6 ‐ 6 ‐ 3 ‐ 3 ‐ 2 ‐ 2 1 1 0 0 0 0 0 – Conjugation (direct transfer between Bacteria) – Transformation by naturally competent Bacteria Bas E. Dutilh – Transduction by bacteriophages Systems Biology: Bioinformatic Data Analysis Utrecht University, March 23 rd 2015 – HGT not just in Bacteria! Pairwise sequence alignments Align GCCCTAGCG to GCGCAATG . A C G T 1 A • What is the optimal alignment? C ‐ 1 1 ‐ 1 ‐ 1 1 G – Many solutions are possible T ‐ 1 ‐ 1 ‐ 1 1 • The most fundamental operation in bioinformatics, used Gap penalty: ‐ 2 • Depends on substitution matrix and gap penalty to identify sequence homology – You could calculate alignment scores for all possible alignments: – (Homologous: similarity by descent from common ancestor) • Definition of sequence alignment 1 + 1 – 1 + 1 – 1 + 1 – 1 – 1 – 2 = ‐ 2 – Given two sequences: seqX = X 1 X 2 …X M M seqY = Y 1 Y 2 …Y N – 2 – 1 + 1 – 1 – 1 + 1 – 1 – 1 + 1 = ‐ 4 an alignment is an assignment of gaps to positions 0, …, M in x, and 0, …, N in seqY, so as to line up each letter in one 1 + 1 – 1 + 1 – 1 + 1 – 2 – 1 + 1 = 0 sequence with either a letter or a gap in the other sequence: 1 + 1 – 1 + 1 – 2 – 2 + 1 – 2 – 1 + 1 = ‐ 3 - AG G CTATCAC CT GACC T C CA GG C CGA -- TGCCC --- AGGCTATCACCTGACCTCCAGGCCGATGCCC Etcetera … T AG - CTATCAC -- GACC G C -- GG T CGA TT TGCCC GAC TAGCTATCACGACCGCGGTCGATTTGCCCGAC The optimal alignment Algorithm • A step ‐ by ‐ step set of operations used for: • The optimal alignment maximizes the alignment score – Complex calcula � ons → • We assume that in the optimal alignment of homologous – Data processing sequences: – Automated reasoning – Aligned amino acids or nucleotides are derived from the same – Cooking → amino acids or nucleotides in the ancestor – Thus, an alignment allows us to identify which mutations occurred during evolution • It is not trivial to make sequence alignments • Algorithms can range from simple – The alignment should be reliable to very complex – The method of obtaining the alignment should be reproducible Ab ū ‘Abdall ā h Mu ḥ ammad Ab ū ‘Abdall ā h Mu ḥ ammad ibn M ū s ā al ‐ Khw ā rizm ī ibn M ū s ā al ‐ Khw ā rizm ī – Thus, we use algorithms to make sequence alignments 780 ‐ 850 (Islamic Golden Age) 780 ‐ 850 (Islamic Golden Age) Persian mathematician, Persian mathematician, astronomer, and geographer astronomer, and geographer 1
24 ‐ Mar ‐ 15 Algorithms in bioinformatics Global and local sequence alignments • In biology, algorithms are critical for reproducible data • Pairwise sequence alignment analysis – Line up two sequences to achieve maximal levels of conservation – To assess the degree of similarity and possibility of homology • Algorithms often come in the form of a computer program or script • Are sequences completely or partially homologous? • When writing a scientific article or report: – Programs and program versions should always be cited • Global alignment Global alignment • Citations include reference to the publication, manufacturer, or website • Citations include reference to the publication manufacturer or website – Aligns two sequences from end to end – Full homologs, e.g. resulting from gene duplication • Local alignment – Finds the optimal sub ‐ alignment within two sequences – Custom ‐ made computer scripts should be provided as supplemental material – Partial homologs, e.g. resulting from domain rearrangement Global alignment Possible alignments A C G T A C G T 1 1 A A • Needleman ‐ Wunsch algorithm • Three global alignments are possible C ‐ 1 1 C ‐ 1 1 ‐ 1 ‐ 1 1 ‐ 1 ‐ 1 1 G G – Also known as “dynamic programming” – All three alignments are valid! T ‐ 1 ‐ 1 ‐ 1 1 T ‐ 1 ‐ 1 ‐ 1 1 – Horizontal step: gap in the ver � cal sequence → penalty Gap penalty: ‐ 2 – Ver � cal step: gap in the horizontal sequence → penalty – Diagonal step: residues are aligned – Backtrack from last cell G C C C T A G C G C G 0 0 ‐ 2 ‐ 2 ‐ 4 ‐ 4 ‐ 6 ‐ 6 ‐ 8 ‐ 8 ‐ 10 ‐ 10 ‐ 12 ‐ 12 ‐ 14 ‐ 14 ‐ 16 ‐ 16 ‐ 18 ‐ 18 ‐ 2 ‐ 2 0 0 ‐ 2 ‐ 4 ‐ 4 ‐ 2 G ‐ 2 ‐ 2 1 1 ‐ 1 ‐ 1 ‐ 3 ‐ 3 ‐ 5 ‐ 5 ‐ 7 ‐ 7 ‐ 9 ‐ 9 ‐ 11 ‐ 11 ‐ 13 ‐ 13 ‐ 15 ‐ 15 G G ‐ 2 1 ‐ 1 1 C ‐ 4 ‐ 4 ‐ 1 ‐ 1 2 2 0 0 ‐ 2 ‐ 2 ‐ 4 ‐ 4 ‐ 6 ‐ 6 ‐ 8 ‐ 8 ‐ 10 ‐ 10 ‐ 12 ‐ 12 G ‐ 6 ‐ 6 ‐ 3 ‐ 3 0 0 1 1 ‐ 1 ‐ 1 ‐ 3 ‐ 3 ‐ 5 ‐ 5 ‐ 5 ‐ 5 ‐ 7 ‐ 7 ‐ 9 ‐ 9 1 ‐ 2 = ‐ 1 ‐ 2 ‐ 2 = ‐ 4 C ‐ 8 ‐ 8 ‐ 5 ‐ 5 ‐ 2 ‐ 2 1 1 2 2 0 0 ‐ 2 ‐ 2 ‐ 4 ‐ 4 ‐ 4 ‐ 4 ‐ 6 ‐ 6 • The alignment scores are identical: A ‐ 10 ‐ 10 ‐ 7 ‐ 7 ‐ 4 ‐ 4 ‐ 1 ‐ 1 0 0 1 1 1 1 ‐ 1 ‐ 1 ‐ 3 ‐ 3 ‐ 5 ‐ 5 ‐ 4 ‐ 2 = ‐ 6 ‐ 2 ‐ 2 = ‐ 4 1+1 ‐ 1+1 ‐ 1+1 ‐ 2 ‐ 1+1=0 1+1 ‐ 1+1 ‐ 1+1 ‐ 1 ‐ 2+1=0 1+1 ‐ 1+1 ‐ 2+1 ‐ 1 ‐ 1+1=0 A ‐ 12 ‐ 12 ‐ 9 ‐ 9 ‐ 6 ‐ 6 ‐ 3 ‐ 3 ‐ 2 ‐ 2 ‐ 1 ‐ 1 2 2 0 0 ‐ 2 ‐ 2 ‐ 4 ‐ 4 • Alignments strongly depend on the substitution matrix! T ‐ 14 ‐ 14 ‐ 11 ‐ 11 ‐ 8 ‐ 8 ‐ 5 ‐ 5 ‐ 4 ‐ 4 ‐ 1 ‐ 1 0 0 1 1 ‐ 1 ‐ 1 ‐ 3 ‐ 3 ‐ 2 ‐ 1 = ‐ 3 0 + 1 = 1 G ‐ 16 ‐ 16 ‐ 13 ‐ 13 ‐ 10 ‐ 10 ‐ 7 ‐ 7 ‐ 6 ‐ 6 ‐ 3 ‐ 3 ‐ 2 ‐ 2 1 1 0 0 0 0 0 Protein alignments Using protein sequences to improve DNA alignments • Make a global alignment of these two sequences using the • Protein sequence is more informative BLOSUM62 substitution matrix than DNA sequence – CAPT – 20 amino acids versus 4 nucleotides – CFT – Amino acids share biochemical properties Gap penalty: ‐ 11 – The genetic code (or codon table) is C A P T degenerate 0 0 ‐ 11 ‐ 2 ‐ 22 ‐ 4 ‐ 33 ‐ 6 ‐ 44 ‐ 8 • Mutations in the third nucleotide of a codon C C ‐ 11 ‐ 11 ‐ 2 ‐ 2 9 9 1 1 ‐ 2 ‐ 1 ‐ 2 ‐ 1 ‐ 13 ‐ 13 ‐ 3 ‐ 3 ‐ 24 ‐ 24 ‐ 5 ‐ 5 often translate into the same amino acid F ‐ 22 ‐ 4 ‐ 1 ‐ 2 7 2 ‐ 4 0 ‐ 15 ‐ 2 • These are called synonymous mutations T ‐ 33 ‐ 6 ‐ 13 ‐ 3 ‐ 2 0 6 1 ‐ 1 1 1 • Protein sequences are more conserved in evolution – Allow you to “look back” further in time • DNA sequences can be translated to protein, and then aligned in “protein space” (Note: different color schemes exist that highlight different properties of amino acids, more about this tomorrow) 2
Recommend
More recommend