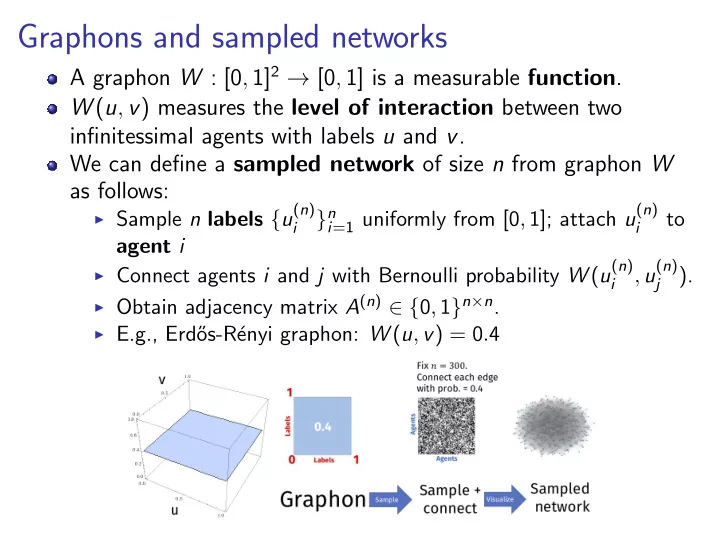

Graphons and sampled networks A graphon W : [0 , 1] 2 → [0 , 1] is a measurable function . W ( u , v ) measures the level of interaction between two infinitessimal agents with labels u and v . We can define a sampled network of size n from graphon W as follows: I Sample n labels { u ( n ) i =1 uniformly from [0 , 1]; attach u ( n ) } n to i i agent i I Connect agents i and j with Bernoulli probability W ( u ( n ) , u ( n ) ). i j I Obtain adjacency matrix A ( n ) ∈ { 0 , 1 } n × n . I E.g., Erd˝ os-R´ enyi graphon: W ( u , v ) = 0 . 4
Contagion in graphons Initial seed set : C 0 ⊆ [0 , 1] (measurable). Threshold function : τ : [0 , 1] → [0 , 1] (piecewise continuous). Set of infected labels in period t : C t Label u is exposed if R 1 0 W ( u , v ) 1 C t − 1 ( v ) dv > τ ( u ) . R 1 0 W ( u , v ) dv . Label u is added to C t if either u ∈ C t − 1 or if u is exposed. In a sampled network : standard linear threshold contagion (Granovetter’78/Morris’00/Kempe-Kleinberg-Tardos’03)
Main theoretical result Given a graphon W , threshold function τ , and initial seed set C 0 , can we predict the terminal set C ∞ of (non-)infected agents in the sampled network? Theorem (informal) In a large enough sampled network, the terminal set of agents (not) infected in a sampled network can be reconstructed with high probability from the terminal set of labels (not) infected in the graphon.
Stochastic Block Model C 0 = [0 , 0 . 1] , τ ( u ) = 0 . 16
Recommend
More recommend