
Multivariate Fundamentals: Rotation Pre-determined Groups Discriminant Analysis aka. Discriminant Function Analysis
Discriminant Analysis (DISCRIM) Analysis for pre-determined groups Objective - Rotate that data so that variation between groups is maximized i.e. “reduce complexity” Different question compared to to PCA & FA (e.g. maximize variation explained) Essentially don’t care how much variance is explained by groups Think of DISCRIM as: “ How far can I separate known groups given measurements of several variables on individuals within these groups ” “ What distinguishes my groups? ” NOT average group measurements Measurements on individuals within the pre-determined groups Sir Ronald Fisher (1890-1962)
How DISCRIM works 1. Find the axis that gives the greatest separation between 2 groups 2. Fix that axis 3. Rotate around the fixed axis to maximize difference between first 2 groups and the 3 rd group 4. Repeat steps 2 & 3 for all groups included y y x ’ x ’ y’ y’ ∝ ∝ x x
The math behind DISCRIM Discriminant functions (DF) are the linear combinations of the original variables Create a DF for every observation in the dataset (like PCA scores) For each function: Column vectors of original variables Linear discriminant (column vector) DF 1 = aX 1 + bX 2 + … + zX n a , b,… z Coefficients for linear model Use matrix algebra to solve the series of discriminant functions for coefficients DF values are what we plot to visualize our analysis
DISCRIM in R Need to install MASS package to run discriminant analysis Dataset for analysis Either MASS or candisk DISCRIM in R: DISCRIM in R: library(MASS) library(candisc) out1=lda(Groups~., data) (MASS package) x=lm(cbind(predictors)~Groups, data) out2=candisc(x, term =“Groups") Column of pre-determined groups You need to define which variables to include in the analysis • If . is specified than all variables in the dataset are include • Else you could include an equation ( e.g. GROUPS~TEMP+PREC+DRY) For candisc you first need to generate a linear regression model of predictors with Group variable as your response variable (function lm), then run candisc for DISCRIM
DISCRIM in R – MASS package output The initial probability of belonging to a group (more important for predicting class) Mean observation values for variables in each pre-defined group Coefficients of linear discriminants are the solutions to our linear functions Proportion of variance explained by linear discriminants MASS will only display solutions for the most significant linear discriminants Discriminants that explain very small portion of the variance are removed
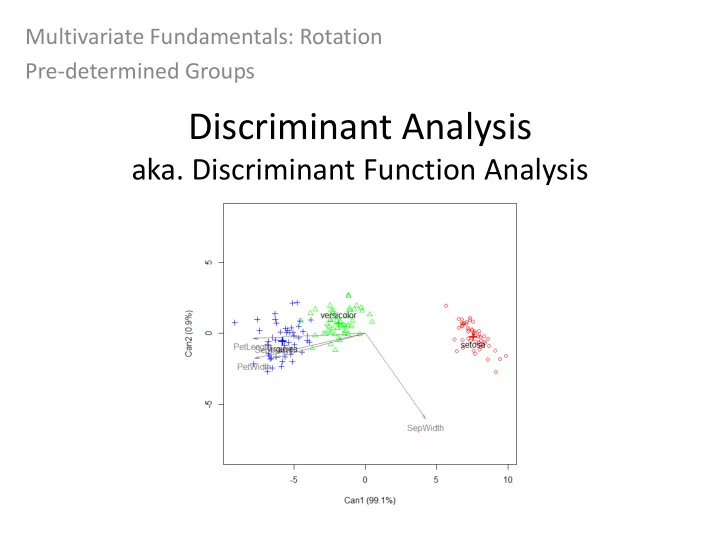
DISCRIM in R – candisc package output Proportion of variance explained by linear discriminants Mean discriminant values for each pre- defined group Standard error of the means are also given By querying the analysis structure we can see the discriminant loadings which tell us the relationship between the DF values and the original variables (like PCA) Again candisc will only display solutions for discriminants that explain the most variation Less information is displayed in the candisc output, but you can get the loadings which are important! Candisc also produces a nicer plot (you will be able to compare outputs in Lab 4)
Using DISCRIM to predict which group Problem : A new skull is found but we don’t know whether it belongs to homo erectus or homo habilis or if it’s a new group? Homo erectus Skull measurement Homo habilis Group centroid New find (unknown origin) Popular method in taxonomy and anthropology How predictions work: 1. Calculate group centroid 2. Find out which centroid is the closest position to the unknown data point New groups are defined when we find a significant difference between new find and predefined groups
Recommend
More recommend