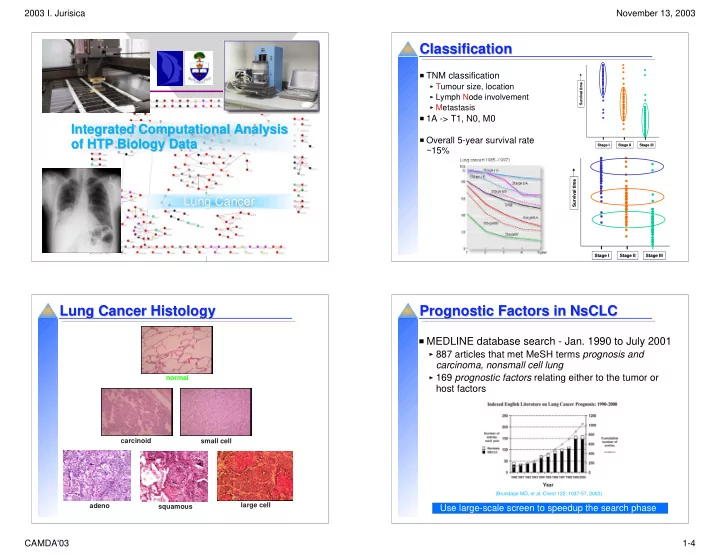

2003 I. Jurisica November 13, 2003 Classification Classification TNM classification Survival time Survival time Tumour size, location Lymph Node involvement Metastasis 1A -> T1, N0, M0 Integrated Computational Analysis Integrated Computational Analysis Overall 5-year survival rate of HTP Biology Data of HTP Biology Data Stage I Stage I Stage II Stage II Stage III Stage III ~15% Survival time Survival time Lung Cancer Lung Cancer Stage I Stage I Stage II Stage II Stage III Stage III Lung Cancer Histology Prognostic Factors in NsCLC Lung Cancer Histology Prognostic Factors in NsCLC MEDLINE database search - Jan. 1990 to July 2001 887 articles that met MeSH terms prognosis and carcinoma, nonsmall cell lung 169 prognostic factors relating either to the tumor or normal host factors carcinoid small cell (Brundage MD, et al, Chest 122: 1037-57, 2002) large cell adeno squamous Use large-scale screen to speedup the search phase CAMDA'03 1-4
2003 I. Jurisica November 13, 2003 Lung Cancer MA Data Sets Overlapping Markers Lung Cancer MA Data Sets Overlapping Markers Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals Histology overlaps - higher distinct adenocarcinoma subclasses. PNAS 98: 13790-95, 2001. Garber ME, et al. Diversity of gene expression in adenocarcinoma of lung. PNAS 98: 13784-89, 2001. well defined, gene set biased before the analysis Wigle DA, et al. Molecular profiling of non-small cell lung cancer and correlation with disease-free survival. Cancer Res 62: 3005-08, 2002. Beer DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nature Outcome overlaps - low Med 8: 816-24, 2002. different array platforms Miura K, et al. Laser capture microdissection and microarray expression analysis of lung adenocarcinoma reveals tobacco-smoking- and prognosis-related molecular profiles. Cancer Res 62: different pools of samples 3244-50, 2002. Sugita M, et al. Combined use of oligonucleotide and tissue microarrays identified cancer/testis mix of histologies, stage, ... antigens as biomarkers in lung carcinoma. Cancer Res 62: 3971-79, 2002. Heighway J, et al. Expression profiling of primary non-small cell lung cancer for target identification. normal vs cancers Oncogene 21: 7749-63, 2002. Wikman H, et al. Identification of differentially expressed genes in pulmonary adenocarcinoma by cancers vs cell lines using cDNA array. Oncogene 21: 5804-13, 2002. cancers vs cancers Virtanen C, et al. Integrated classification of lung tumors and cell lines by expression profiling. PNAS 99: 12357-62, 2002. different analysis biases Pedersen N, et al. Transcriptional gene expression profiling of small cell lung cancer cells. Cancer Res 63: 1943-53, 2003. wide screen first - but then validating known Nakamura H, et al. cDNA microarray analysis of gene expression in pathologic stage IA nonsmall cell lung carcinomas. Cancer 97: 2798-805, 2003. Yamagata N et al. A Training-Testing Approach to the Molecular Classification of Resected Non-Small Cell Lung Cancer, Clinical Cancer Research 9:4695–4704, 2003. Goals Lung Cancer Data Sets Goals Lung Cancer Data Sets ADC SCC LC Tumor Normal Cell line Array Pool of Bhattacharjee 127 21 26 17 U95 12.6K Genes Garber 41 16 5 5 5 SA 24K Wigle 19 14 16 SA 19K Beer 86 10 UGFL 7K Pool of Miura 19 SA 18.4K Samples Sugita 4 U95 12.6K Heighway 39 33 SA 47K Wikman 14 4 HCG 1.2K Set of Markers Set of Markers Virtanen 44 5 41 SA 7.7K Pedersen 18 21 U95 12.6K Nakamura 10 10 425 Yamagata 9 11 8 3 6 SA 5.2K CAMDA03 data sets overlap 2449 5 data sets overlap 1639 Biases any two sets 5885 out of 338 Stanford genes 55 in 5 sets; 75 in CAMDA Data Results Discover something new and useful CAMDA'03 5-8
2003 I. Jurisica November 13, 2003 OCI: BTSVQ Clustering of Cases based on PCR Values OCI: BTSVQ Clustering of Cases based on PCR Values Stanford Stanford Microarrays Microarrays Genes = 11 Genes = 11 Cases =39 Cases = 131 bad ok Early filtering good 23,100 (17,108) genes -> 918 (835) 1.0 1.0 No overall trend with survival 0.8 0.8 Adeno group can be further partitioned Disease-free Survival Disease-free Survival 0.6 0.6 0.4 0.4 Log rank p-value = 0.23 Log rank p-value = 0.23 0.2 0.2 1 , n= 47 1 , n= 47 2 , n= 84 2 , n= 84 0.0 0.0 No single gene may cut it, but a panel of them can be predictive 0 0 1 1 2 2 3 3 4 4 5 5 6 6 Years to first failure Years to first failure Harvard/Michigan Definition Harvard/Michigan Definition Microarrays Microarrays Harvard Michigan Paralysis of analysis U95 -> 3,312 genes -> UGFL -> 4,966 genes too many hypothesis to follow 675 genes some clusters have too many exceptions some clusters are highly significant survival trend statistical significance may not imply biological relevance differential, and show survival trend Intellectual prosthesis managing knowledge differential analysis Design reasoning biology understanding Remembering Data hypothesis generation Retrieving Reasoning Interpretation Analysis CAMDA'03 9-12
2003 I. Jurisica November 13, 2003 Microarray Data Analysis & Visualization Aims Microarray Data Analysis & Visualization Aims Microarrays Microarrays From hypothesis-driven research to hypothesis-generation Cluster Gene From biology understanding to individualized Selection medicine or from information-based, individualized medicine to understanding SOM & k-means Move from static to "dynamic" analysis Novel Approach to MA Clustering & Visualization Novel Approach to MA Clustering & Visualization Analysis: MA Analysis: MA Microarrays Microarrays 2-way approach unbiased clustering of both samples and genes Top down, iterative k-means clustering SOM Vector quantization Vector projection Stat significance CAMDA'03 13-16
2003 I. Jurisica November 13, 2003 Recurrence/Survival Recurrence/Survival Harvard Stanford Microarrays Microarrays 12K genes 22696 genes 6/7 category 1 7/11 category 0 alive Same histology dead Same stage Lung cancer Different Pattern Michigan ~90% of the - stage I, II, III 12K genes samples show a Different histology Different stage strong pattern Same Pattern Histology Conclusion on MA Histology Conclusion on MA Harvard coid-coid Microarrays Microarrays Large number of samples more homogenous study of subtypes with detailed outcome information stronger validation Validation in the clinical context different method different samples different method & different samples Knowledge discovery in the biological context biology of tumorigenesis Use OCI CAMDA'03 17-20
2003 I. Jurisica November 13, 2003 Integrated Analysis PPI Data Analysis Integrated Analysis PPI Data Analysis Protein-Protein Interactions Protein-Protein Interactions Prediction Protein Protein Microarrays Microarrays Interactions Interactions Interpretation Interpretation - noise - unknowns, uncharacterized degree, hubs, articulation points, siblings - system context pathways, complexes function Structure - function relationship PPI Data Analysis Analysis: PPI PPI Data Analysis Analysis: PPI Top 3% Degree 1 Protein-Protein Interactions Transcription 2x Translation 12x 4x Uncharacterized Metabolism Hubs&Art.pnts Lethals G.maintenance 2x Protein fate Cellular org. - 80% of art. points in DDR - lethal - 92% of art. points in MAPK - lethal CAMDA'03 21-24
Recommend
More recommend