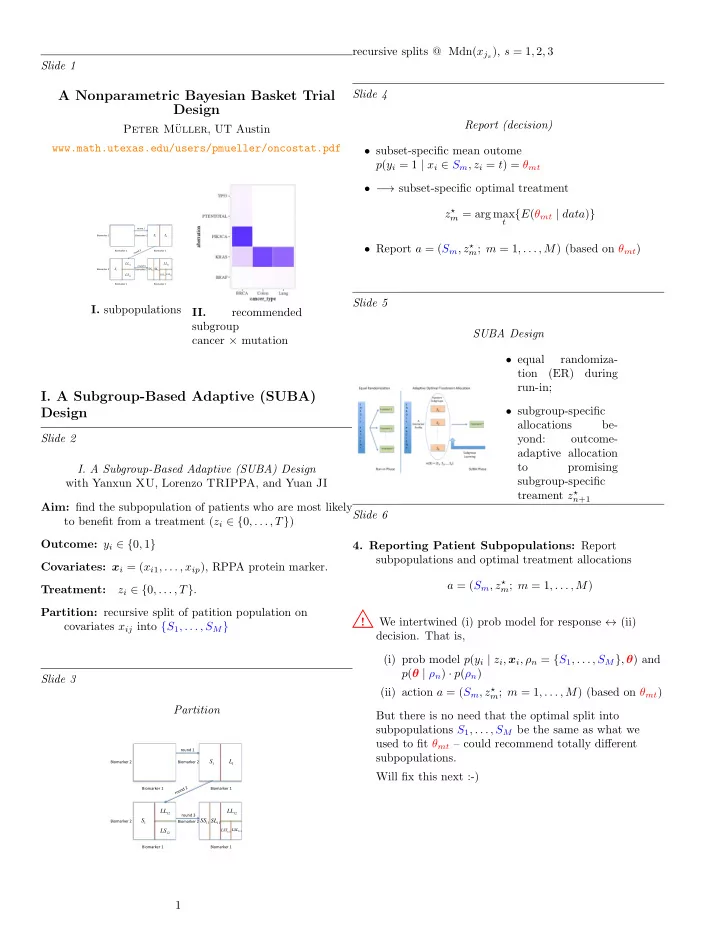

recursive splits @ Mdn( x j s ), s = 1 , 2 , 3 Slide 1 A Nonparametric Bayesian Basket Trial Slide 4 Design Report (decision) Peter M¨ uller , UT Austin www.math.utexas.edu/users/pmueller/oncostat.pdf • subset-specific mean outome p ( y i = 1 | x i ∈ S m , z i = t ) = θ mt • − → subset-specific optimal treatment z ⋆ m = arg max t { E ( θ mt | data ) } ¡ ¡ ¡round ¡1 ¡ Biomarker ¡2 ¡ Biomarker ¡2 ¡ S 1 L 1 • Report a = ( S m , z ⋆ m ; m = 1 , . . . , M ) (based on θ mt ) Biomarker ¡1 ¡ Biomarker ¡1 ¡ LL 12 LL 12 ¡ ¡ ¡ ¡round ¡3 ¡ Biomarker ¡2 ¡ S 1 Biomarker ¡2 ¡ SS 11 SL 11 LS 12 LSS 121 LSL 121 Biomarker ¡1 ¡ Biomarker ¡1 ¡ Slide 5 I. subpopulations II. recommended subgroup SUBA Design cancer × mutation • equal randomiza- tion (ER) during run-in; I. A Subgroup-Based Adaptive (SUBA) • subgroup-specific Design allocations be- Slide 2 yond: outcome- adaptive allocation to promising I. A Subgroup-Based Adaptive (SUBA) Design subgroup-specific with Yanxun XU , Lorenzo TRIPPA , and Yuan JI treament z ⋆ n +1 Aim: find the subpopulation of patients who are most likely Slide 6 to benefit from a treatment ( z i ∈ { 0 , . . . , T } ) Outcome: y i ∈ { 0 , 1 } 4. Reporting Patient Subpopulations: Report subpopulations and optimal treatment allocations Covariates: x i = ( x i 1 , . . . , x ip ), RPPA protein marker. a = ( S m , z ⋆ m ; m = 1 , . . . , M ) Treatment: z i ∈ { 0 , . . . , T } . Partition: recursive split of patition population on ! We intertwined (i) prob model for response ↔ (ii) covariates x ij into { S 1 , . . . , S M } decision. That is, (i) prob model p ( y i | z i , x i , ρ n = { S 1 , . . . , S M } , θ ) and p ( θ | ρ n ) · p ( ρ n ) Slide 3 (ii) action a = ( S m , z ⋆ m ; m = 1 , . . . , M ) (based on θ mt ) Partition But there is no need that the optimal split into subpopulations S 1 , . . . , S M be the same as what we used to fit θ mt – could recommend totally different ¡ ¡ ¡round ¡1 ¡ subpopulations. S 1 L 1 Biomarker ¡2 ¡ Biomarker ¡2 ¡ Will fix this next :-) Biomarker ¡1 ¡ Biomarker ¡1 ¡ LL 12 LL 12 ¡ ¡ ¡ ¡round ¡3 ¡ Biomarker ¡2 ¡ S 1 Biomarker ¡2 ¡ SS 11 SL 11 LS 12 LSL 121 LSS 121 Biomarker ¡1 ¡ Biomarker ¡1 ¡ 1
II. IMPACT II: a basket trial for targeted Utility: we favor a subpopulation with difference (relative therapy to the overall population) in log hazards ratio (HR), large size and parsimonious description with few Slide 7 covariates, n ( a ) α II. IMPACT II: a basket trial for targeted therapy u ( a , θ ) = (HR( a, θ ) − β ) · ( | I | + 1) γ with Yanxun Xu , Don Berry and A. Tsimboridou with n ( a ) = size of the subpopulation, and θ indexes the sampling model, � Bayes rule: Report a ⋆ = arg max a u ( a , θ ) dp ( θ | data ) , Trial: study of targeted agents in metastatic cancers. � �� � U ( a ) Patients: metastatic cancer (thyroid, ovarian, melanoma, maximizing expected utility U ( a ). lung . . . ) Alternative utility: Foster, Taylor & Ruberg (2011, Treatments: therapy that targets particular molecular StatMed) use aberrations (TT) vs. standard of care (S) Q ( A ) = enhanced treatment effect − average trt effect Population finding: y i = PFS; heterogeneous pat and sensitivity and specificity to evaluate a reported population – subpopulation A . x i = different mutations; different cancers; baseline covs . . . Model: Decicsion problem and solution remain meaningful Treatment might be effective for a subgroup across for any model, e.g. a nonparametric Bayes model diseases (“always right”) Strategy (for pop. finding): similar to (I) , but separate (i) prob model p ( y | x , θ ), vs. (ii) decision problem a ∈ A . 2 Simulation Slide 10 1 Decision problem 4. Simulation Slide 8 Scenarios: 7 scenarios, p = 8 covariates (7 mutations, 1 cancer type). 2. Decision Problem Simulation truth is a log normal regression for y i ∈ ℜ . Actions: Report a (i.e., one ) subgroup of patients who True subgroups: Evaluation of (frequentist) error rates might most benefit from TT: requires “true” subgroups. � Defined as winner under U 0 ( a ) = u ( a, θ ) dp 0 ( θ ) under a : patients with x ij k = x ⋆ j k , k = 1 , . . . , K a “true” assumed sampling model p 0 . select (multiple) combination of mutations and cancer • Evaluate u ( a, · ) under the simulation truth using the true log hazards ratios for a subgroup a . Prob model: � (indexed by θ ) • Repeat for all poss subgroups. Decision problem: separate inference (predicting PFS) vs. • The top subgroup is labeled as “truth” decision (report subpopulation). Results: next slide. • no need for multiplicity control • arbitrary prob model • disentagle stat significance vs. clinical relevance • allow for variable # covs. Slide 9 2
Slide 14 Slide 11 Comparison with simple designs Matching cancer types and mutations Difference between estimated and true TE NAIVE INDEP OURS value 0.5 mutation PTEN mutation PTEN mutation PTEN PIK3CA PIK3CA PIK3CA 0.4 TP53 TP53 Sc 3: MET MET MET 0.3 FGFR FGFR FGFR BRAF BRAF BRAF 0.2 value BRCA Ovary Lung BRCA Ovary Lung BRCA Ovary Lung 0.1 PTENTOTAL PTENTOTAL 1.00 tumor_type tumor_type tumor_type 0.0 mutation mutation 0.75 NAIVE INDEP OURS value PIK3CA PIK3CA 0.5 mutation PTEN mutation PTEN mutation PTEN 0.4 0.50 PIK3CA PIK3CA PIK3CA Sc 4: MET MET MET 0.3 FGFR FGFR FGFR KRAS KRAS 0.25 BRAF BRAF BRAF 0.2 BRCA Ovary Lung BRCA Ovary Lung BRCA Ovary Lung 0.1 tumor_type tumor_type tumor_type 0.00 0.0 BRAF BRAF NAIVE SEPARATE OURS value 0.5 mutation PTEN mutation PTEN mutation PTEN BRCA Lung Colon BRCA Lung Colon 0.4 PIK3CA PIK3CA PIK3CA tumor_type tumor_type Sc 5: MET MET MET 0.3 FGFR FGFR FGFR BRAF BRAF BRAF 0.2 BRCA Ovary Lung BRCA Ovary Lung BRCA Ovary Lung 0.1 tumor_type tumor_type tumor_type 0.0 Massive repeat simulation under a hypothetical truth (left), NAIVE SEPARATE OURS value 0.5 and frequency (under repeat simulation) of recommending mutation PTEN mutation PTEN mutation PTEN 0.4 PIK3CA PIK3CA PIK3CA Sc 6: MET MET MET mutation-tumor pairs a ⋆ with treatment effect different from 0.3 FGFR FGFR FGFR BRAF BRAF BRAF 0.2 BRCA Ovary Lung BRCA Ovary Lung BRCA Ovary Lung 0.1 tumor_type tumor_type tumor_type the overall population (right) 0.0 NAIVE= one common TE; SEPARATE= separate studies Slide 12 Slide 15 Operating Characteristics: Error Rates TIE = p ( H c 0 | H 0 ) type-I error FNR = p ( H 0 | H c 0 ) false neg. rate Summary TPR = p ( H 1 | H 1 ) true positive r. FSR = p ( H a | H c a ) false subgroup r. TSR = p ( H a | H a ) true subgroup r. FPR = p ( H 1 | H a ) false positive r. Basket trial design: A Bayesian approach with a sensible strategy to detect winning subgroups: Truth Subgroup Effect • Separate model fit vs. report of optimal Decision H 0 H a H 1 subpopulation. H 0 1-TIE FNR • Coherent posterior probabilites for subgroup H a TSR a FSR effects. H 1 FPR a TPR • Multiplicity control is achieved by choice of priors • Choose α, β, γ to control TIE and (average) FSR. & by controlling frequentist error rate. • All but the TIE require additional specification: – effect size for FNR, TPR and FSR. Xu, Trippa, Mueller, Ji (2016 Stat in Biosc), “Subgroup-Based Adaptive (SUBA) Designs for Multi-Arm – TSR and FPR depend on specific subgroup a . Biomarker Trials,” uller, Tsimberidou, Berry (2018 Biom J). A Xu, M¨ population-finding design with covariate-dependent random partitions Slide 13 Simulation results Scenario TIE TSR TPR FSR FNR FPR 1 0.05 - - - - - 2 - - .90 - - - 3 - .88 - .04 .10 .00 4 - .66 - .05 .04 .00 5 - .77 - .03 .01 .00 6 - .78 - .02 .03 .00 ⋆ scenario 1 is true H 0 all others are true subgroup and overall effects TIE = p ( H c 0 | H 0 ) type-I error FNR = p ( H 0 | H 1 ) false negative rate TPR = p ( H 1 | H 1 ) true positive r. FSR = p ( H a | H c a ) false subgroup r. TSR = p ( H a | H a ) true subgroup r. FPR = p ( H 1 | H a ) false positive r. 3
Recommend
More recommend