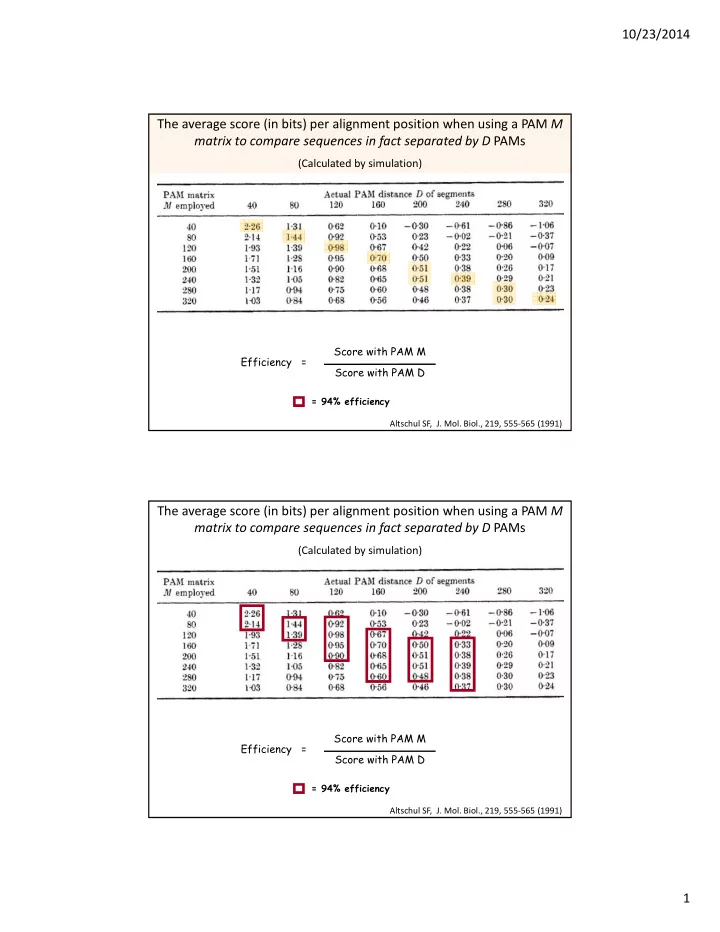

10/23/2014 The average score (in bits) per alignment position when using a PAM M matrix to compare sequences in fact separated by D PAMs (Calculated by simulation) Score with PAM M Efficiency = Score with PAM D = 94% efficiency Altschul SF, J. Mol. Biol., 219, 555 ‐ 565 (1991) The average score (in bits) per alignment position when using a PAM M matrix to compare sequences in fact separated by D PAMs (Calculated by simulation) Score with PAM M Efficiency = Score with PAM D = 94% efficiency Altschul SF, J. Mol. Biol., 219, 555 ‐ 565 (1991) 1
10/23/2014 The relative entropy of a substitution matrix is given in bits per position and can be calculated from S[] using the equations n S [ i , j ] n n H q S i j n [ , ] q p p e and ij ij i j i , j BLOSUM PAM Sequence identity bits/site bits/site 20 2.95 83% 30 2.57 60 2.00 63% 70 1.60 90 1.18 100 1.18 43% 80 0.99 120 0.98 38% 60 0.66 160 0.70 30% 50 0.52 200 0.51 25% 45 0.38 250 0.36 20% The “Twilight” Zone • The scale indicates % identity in local % Identity alignments (MSPs). 100 • The Twilight Zone – Around 20% ‐ 35% identity 75 Pairwise searches – Difficult to distinguish between MSPs in related sequences and “chance” alignments 50 PSSM/HMM Twilight zone 25 searches Structure prediction 0 2
10/23/2014 First in ‐ class exam scores 100.00 Mean: 84.8 95.00 90.00 85.00 80.00 75.00 70.00 3
Recommend
More recommend