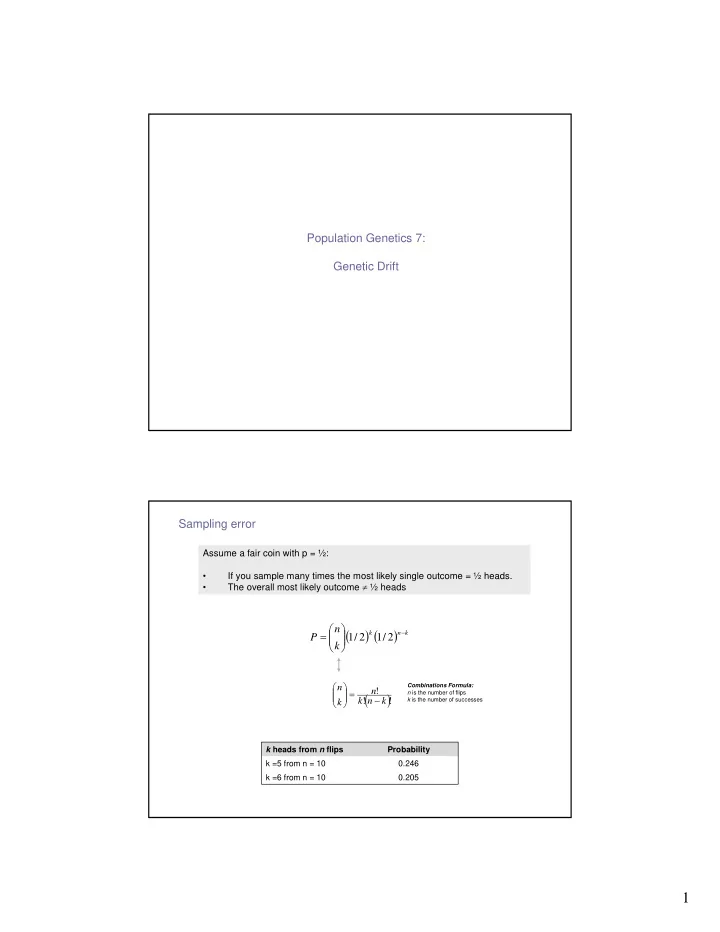

Population Genetics 7: Genetic Drift Sampling error Assume a fair coin with p = ½: • If you sample many times the most likely single outcome = ½ heads. The overall most likely outcome ≠ ½ heads • ⎛ ⎞ n ( ) ( ) − = ⎜ ⎟ k n k P 1 / 2 1 / 2 ⎜ ⎟ ⎝ ⎠ k ⎛ ⎞ Combinations Formula: n ⎜ ⎟ = n ! ( ) n is the number of flips ⎜ ⎟ − ⎝ ⎠ k ! n k ! k is the number of successes k k heads from n flips Probability k =5 from n = 10 0.246 k =6 from n = 10 0.205 1
Sampling error The long term average value for p is 0.5; let’s call that E( p ). How do we improve our changes of getting something close E( p )? N flips p <0.35 p = 0.35-0.45 p = 0.45-0.55 p = 0.55-0.65 p <0.65 variance 10 0.16 0.21 0.25 0.21 0.16 0.025 20 0.06 0.19 0.50 0.19 0.06 0.0125 50 0.002 0.16 0.68 0.16 0.002 0.005 If we flipped the coin 1000 times: we get very close to E( p ) in a single try, but not exactly. Genetic drift Consider a diploid population: • Ideal population: no sampling errors because infinite population size • Natural population: finite size and finite sample of gametes [errors] Example: Let’s assume: A = p = 0.75; a = q = 0.25; N = 500 This generation: 200 individuals reproduce [400 gametes] This is a binomial sampling problem: The probability of getting p = 0.75 and q = 0.25 in next generation is: ⎛ ⎞ 400 ( ) ( ) = ⎜ ⎟ 300 100 P 0 . 75 0 . 25 ⎜ ⎟ ⎝ 300 ⎠ P = 0.046 2
Genetic drift Generation 0 Generation 0 Generation 1 Generation 1 Generation 2 Generation 2 Generation 3 Generation 3 Draw Draw Draw Draw Draw Draw 4 : 6 4 : 6 7 : 3 7 : 3 8 :2 8 :2 Restock Restock Restock Restock Restock Restock white = 0.5 white = 0.5 white = 0.4 white = 0.4 white = 0.7 white = 0.7 white = 0.8 white = 0.8 Genetic drift is the accumulation of random sampling fluctuations in allele frequencies over generations. Genetic drift The magnitude of change in allele frequencies is inversely proportional to the sample size: Ideal population with finite size 1 and finite gamete sample per N generation. See last slide for example Remember that natural populations are less than ideal in many more ways! In most natural populations the effective size (N e ) will be less than the census size. The magnitude of drift in natural populations is: 1 N e Drift and inbreeding effects are not independent! 3
Genetic drift N e = 100 N e = 1000 N e = 10000 N e = 50000 Genetic drift If we run this simulation long enough it will go to fixation of loss; it just takes much longer • rate to fixation [under drift] slows with increasing in N e • ultimate fate is fixation or loss 4
Genetic drift Generation 0 Generation 0 Generation 1 Generation 1 Generation 2 Generation 2 Generation 3 Generation 3 Draw Draw Draw Draw Draw Draw 4 : 6 4 : 6 7 : 3 7 : 3 8 :2 8 :2 Restock Restock Restock Restock Restock Restock white = 0.5 white = 0.5 white = 0.4 white = 0.4 white = 0.7 white = 0.7 white = 0.8 white = 0.8 In natural populations it is N e that counts! Genetic drift What is the fate (on average) of a new mutant? The probability of fixation of a new mutant is its frequency ( p or q ) in the population: 1 N e This is al low as it gets. The fate of most new mutations is LOSS due to drift. W AA = 0.5; W Aa = 0.5; W aa = 1: Probability of fixation actually declines as N e • ideal population: probability of fixation = 1 decreases! • population with N e = 50: probability of fixation ~ 0.25 5
Genetic drift 10 Independent populations; each started with p = q = 0.5 1 1 1 1 1 9 . 0 8 . 0 . 0 8 8 0 . 0 . 8 0 8 . 7 . 0 . 6 0 6 . 0 0 . 6 . 0 6 0 6 . 5 . 0 4 . 0 0 . 4 . 4 0 0 . 4 0 . 4 Allele frequency 0 3 . 0 2 . 2 0 . . 2 0 0 2 . 0 . 2 0 . 1 0 0 0 0 0 1 3 5 7 9 11 13 15 17 19 21 23 25 1 3 5 7 9 11 13 15 17 19 21 23 25 1 3 5 7 9 11 13 15 17 19 21 23 25 1 3 5 7 9 11 13 15 17 19 21 23 25 1 3 5 7 9 11 13 15 17 19 21 23 25 1 1 1 1 1 * 8 . 0 0 . 8 0 8 . 0 8 . . 0 8 0 . 6 0 . 6 0 . 6 0 . 6 0 . 6 0 4 . 0 4 . 4 . 0 0 4 . 4 . 0 2 . 0 0 . 2 0 . 2 0 2 . 2 . 0 * 0 0 0 0 0 1 3 5 7 9 11 13 15 17 19 21 23 25 1 3 5 7 9 11 13 15 17 19 21 23 25 1 3 5 7 9 11 13 15 17 19 21 23 25 1 3 5 7 9 11 13 15 17 19 21 23 25 1 3 5 7 9 11 13 15 17 19 21 23 25 Generation N e = 50; generations = 50 * = fixation Changes in allele frequency due to drift are unpredictable! Note if we ran more generations, more popns would go to fixation Genetic drift 25 45 number of populations 40 20 35 30 15 25 20 10 15 10 5 5 0 0 0 allele frequency 1 1 2 3 4 5 6 7 8 9 10 11 12 13 0 allele frequency 1 1 2 3 4 5 6 7 8 9 10 11 12 13 initial distribution; t = 0 generations distribution after t = 50 generations 6
Genetic drift The effects of drift are cumulative over time. The effects of drift are predictable as averaged over time and populations: 1. loss of variation within populations 2. gain in variation between populations Does genetic drift affects heterozygosity? Let x be the amount of change in p and q in a population due to drift. As we have seen the long term average, E(x), due to drift will be zero because changes in p and q are equally likely to be positive or negative. Given E(x) = 0, what happens to heterozygosity? Does heterozygosity change at all? Let’s start with HW at generation t: H t = 2 pq The allele frequencies, p and q, will change from generation to generation by the amount x: H t+1 = 2( p + x )( q – x ) H t+1 = 2pq + 2x(q – p) – 2x 2 Although E(x) = 0, the expected value of x-squared, E(x 2 ), is always positive. E(2pq + 2x(q – p) – 2x 2 ) 2pq – 2x 2 Heterozygosity is expected reduced by genetic drift. Nice, eh? 7
Genetic drift and inbreeding are not independent 1. Unequal numbers in successive generations ⎡ ⎤ 1 1 1 1 1 1 = + + + + ⎢ ⎥ ... (approx.) ⎢ ⎥ N g N N N N ⎣ ⎦ e 1 2 3 g 2. Different numbers of males and females 1 1 1 = + (approx.) N 4 N 4 N e m f 3. Variance in reproductive success (other than male verse female) − 4 N 2 ( ) = v N e + V 2 k Bottlenecks and founder effects Bottleneck: is a single, extraordinarily large, reduction in population size Pre-bottleneck population Post-bottleneck population Bottleneck event 1. Change in allele frequencies, as compared with pre-bottleneck population 2. Reduction in diversity 8
Bottlenecks and founder effects Effective population size is dominated by historical lows and can be very much lower than current census size. 120,000 Population crash population census size Population recovered to historical high 100,000 80,000 60,000 Ave N 40,000 20,000 N e 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Time ⎡ ⎤ 1 1 1 1 1 1 = + + + + ⎢ ⎥ ... (approx.) ⎢ ⎥ N g ⎣ N N N N ⎦ e 1 2 3 g Dispersal event to a New population Parental population neighbouring island Island 1 Island 2 9
Polydactyly caused by the homozygous recessive disease Ellis-van Creveld syndrome Other symptoms of this disease include dwarfisms, abnormalities of the nails and teeth, and a hole between the two upper chambers of the heart. Picture wing Drosophila Direction of colonization Direction of archipelago growth 10
Two species that have suffered extreme bottlenecks due to commercial harvesting Northern elephant seal Northern right whale Excellent population recovery Poor population recovery Genetic drift Keynotes: • Genetic drift influences allele frequency and genotype frequency. • Drift decreases diversity within populations and increases diversity between populations. • Under drift, fixation rate is determined by N e ; the probability of fixation by p . • In specific cases the outcome of genetic drift is unpredictable. • The effects of drift are predictable as an average over populations. • Drift might reduce a population’s ability to evolve in response to new selective pressures (remember Trudy MacKay’s experiments). Alternatively, some believe that drift could actually increase the rate of speciation. • Drift is particularly important in rare and endangered species. • Founder effects may play an important role in some speciation events 11
HW model: no change in frequencies Alt model; change in frequencies (molecular evolution) Change in frequencies Agency Genotype Allele Notes Linkage no no Creates disequilibrium among loci Inbreeding yes no Acts on all loci in genome; results in loss of heterozygosity Assortative Mating yes no Only acts on the locus subject to assortment, and those loci linked to it Migration a yes yes Depends of migration rate and frequency differences between populations Mutation yes yes Very very very slow Natural Selection yes yes Acts on the locus subject to selection, and those loci linked to it Genetic Drift yes yes Acts on all loci in the genome; results in loss of heterozygosity and loss of alleles 12
Recommend
More recommend