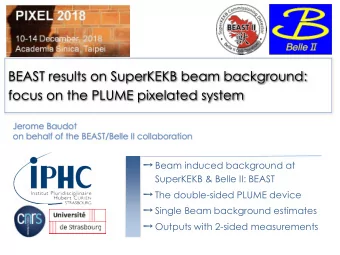
Taming the Beast Workshop Bayesian inference of species tree - PowerPoint PPT Presentation
Taming the Beast Taming the Beast Workshop Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent Bayesian inference of species tree Molecular clock model Felsenstein likelihood and
Taming the Beast Taming the Beast Workshop Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent Bayesian inference of species tree Molecular clock model Felsenstein likelihood and *BEAST Posterior distribution starBEAST2 References Chi Zhang June 28, 2016 1 / 19
Species tree Taming the Beast Bayesian inference of species tree ◮ Species tree — the phylogeny representing the relationships Species & gene trees *BEAST among a group of species Species tree prior Multispecies coalescent Molecular clock model Felsenstein likelihood Posterior distribution starBEAST2 References Figure adapted from [Rogers and Gibbs, 2014] ◮ Gene tree — the phylogeny for sequences at a particular gene locus from those species 2 / 19
Gene tree discordance Taming the Beast Bayesian inference of species tree Species & gene trees ◮ Incomplete lineage sorting *BEAST Species tree prior Multispecies coalescent Molecular clock model Felsenstein likelihood Posterior distribution starBEAST2 References Figure adapted from [Patterson et al., 2006] 3 / 19
Gene tree discordance Taming the Beast Bayesian inference of species tree Species & gene trees ◮ Horizontal gene transfer *BEAST Species tree prior Multispecies coalescent ◮ Gene duplication and loss Molecular clock model Felsenstein likelihood Posterior distribution starBEAST2 References Figure adapted from [Degnan and Rosenberg, 2009] 4 / 19
Gene tree discordance Taming the Beast Bayesian inference of ◮ Hybridization species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent Molecular clock model Felsenstein likelihood Posterior distribution starBEAST2 References Figure adapted from [Li et al., 2016] 5 / 19
Species tree inference and *BEAST Taming the Beast Bayesian inference of species tree Species & gene trees ◮ A Bayesian method to infer species tree from multilocus *BEAST Species tree prior sequence data [Heled and Drummond, 2010] Multispecies coalescent Molecular clock model ◮ *BEAST, a functionality of BEAST2 Felsenstein likelihood Posterior distribution starBEAST2 ◮ Gene trees are embedded References in the species tree under the multispecies coalescent model [Rannala and Yang, 2003] ◮ incomplete lineage sorting ◮ Gene trees are independent among loci Human Gorilla Chimp 6 / 19
Species tree prior Taming the Beast Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent ◮ The prior for species tree S has two parts: Molecular clock model Felsenstein likelihood Posterior distribution P ( S ) = P ( S T ) P ( N ) starBEAST2 References ◮ S T — species time tree ◮ N — population size functions ◮ P ( S T ) — typically a Yule (pure-birth) or birth-death prior ◮ we can assign a hyperprior for the speciation (birth) rate (and extinction (death) rate, if birth-death) ◮ P ( N ) — constant or continuous-linear 7 / 19
Species tree prior Taming the Beast Bayesian inference of species tree Species & gene trees ◮ Constant population sizes *BEAST Species tree prior Multispecies coalescent Molecular clock model Felsenstein likelihood Posterior distribution starBEAST2 References Figure adapted from [Drummond and Bouckaert, 2015] 8 / 19
Species tree prior Taming the Beast Bayesian inference of species tree ◮ Continuous-linear population sizes Species & gene trees *BEAST Species tree prior Multispecies coalescent Molecular clock model Felsenstein likelihood Posterior distribution starBEAST2 References Figure adapted from [Drummond and Bouckaert, 2015] 9 / 19
Species tree prior Taming the Beast Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent Molecular clock model Felsenstein likelihood ◮ In *BEAST, the prior type for N is fixed to gamma Posterior distribution starBEAST2 ◮ The gamma shape parameter k is fixed to 2, but we can References assign a hyperprior for ψ , the scale parameter of the gamma ◮ (This ψ parameter is called ”population mean” in Beauti, but the prior mean is actually 2 ψ when the population sizes are constant) 10 / 19
Multispecies coalescent model Taming the Beast Bayesian inference of ◮ The prior for gene tree g , given species tree S species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent Molecular clock model Felsenstein likelihood Posterior distribution starBEAST2 References Figure adapted from [Drummond and Bouckaert, 2015] 11 / 19
Multispecies coalescent model Taming the Beast Bayesian inference of species tree Species & gene trees ◮ The prob. distribution of gene time tree g given species tree *BEAST Species tree prior S , is: Multispecies coalescent 2 s − 1 Molecular clock model � P ( g | S ) = P ( L j ( g ) | N j ( t )) Felsenstein likelihood Posterior distribution j = 1 starBEAST2 References ◮ s — number of extant species (2 s − 1 branches totally) ◮ N j ( t ) — population size function (linear) ◮ L j ( g ) — coalescent intervals for genealogy g that are contained in the j ’th branch of species tree S 12 / 19
Molecular clock model Taming the Beast Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent ◮ P ( c ) — prior for the molecular clock model of genealogy g Molecular clock model ◮ strict clock — typically fix to 1.0 for the first locus, and Felsenstein likelihood Posterior distribution infer the relative clock rates for the rest loci starBEAST2 ◮ relaxed clock References ◮ P ( θ ) — prior for the substitution model parameters ◮ e.g. HKY85, ◮ Prior for transition/transversion rate ratio ( κ ), e.g. gamma(2,1) ◮ Prior for base frequencies ( π T , π C , π A , π G ), e.g. Dirichlet(1,1,1,1) 13 / 19
Felsenstein likelihood Taming the Beast Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent Molecular clock model Felsenstein likelihood Posterior distribution ◮ The probability (likelihood) of data d i (alignment at locus starBEAST2 References i ), given the gene time tree g i , molecular clock c i , and substitution model θ i , is: P ( d i | g i , c i , θ i ) 14 / 19
Priors and likelihood Taming the Beast Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent Molecular clock model ◮ P ( S ) — prior for species tree Felsenstein likelihood Posterior distribution starBEAST2 ◮ P ( g i | S ) — prior for gene tree i (multispecies coalescent) References ◮ P ( c i ) — prior for clock rate of locus i ◮ P ( θ i ) — prior for substitution parameters of locus i ◮ P ( d i | g i , c i , θ i ) — likelihood of data at locus i 15 / 19
Posterior Taming the Beast Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent Molecular clock model Felsenstein likelihood ◮ The posterior distribution of the species tree S and other Posterior distribution paremeters given data D is: starBEAST2 References n � P ( S , g , c , Θ | D ) ∝ P ( S ) P ( g i | S ) P ( c i ) P ( θ i ) P ( d i | g i , c i , θ i ) i = 1 ◮ The data D = { d 1 , d 2 , . . . , d n } is composed of n alignments, one per locus. 16 / 19
Integrating out population sizes Taming the Beast Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent Molecular clock model ◮ Assume constant population sizes Felsenstein likelihood Posterior distribution ◮ Assign i.i.d inverse-gamma( α , β ) prior for N j starBEAST2 References ◮ mean = β/ ( α − 1 ) ◮ The population sizes N can be integrated out from P ( g | S ) [Jones, 2015] ◮ Specify α and β in the invgamma prior (instead of ψ in the gamma prior) 17 / 19
starBEAST2 Taming the Beast Bayesian inference of species tree Species & gene trees *BEAST Species tree prior Multispecies coalescent ◮ A more efficient implementation and an upgrade of *BEAST Molecular clock model Felsenstein likelihood Posterior distribution starBEAST2 ◮ Population sizes integrated out [Jones, 2015] References ◮ Relaxed molecular clock per species tree branch (instead of per gene tree branch) ◮ More efficient MCMC proposals for the species tree and gene trees (coordinated operators) [Jones, 2015, Rannala and Yang, 2015] ◮ Available at github.com/genomescale/starbeast2 , will be released soon (as a BEAST2 add-on) 18 / 19
Recommend
More recommend
Explore More Topics
Stay informed with curated content and fresh updates.