Robustness of Complex Dynamics in Biology Workshop on Uncertain - PDF document
From qualitative to quantitative models C. Breindl, D. Schittler, S. Waldherr, and F. Allgwer Institute for Systems Theory and Automatic Control Robustness of Complex Dynamics in Biology Workshop on Uncertain Dynamical Systems University of
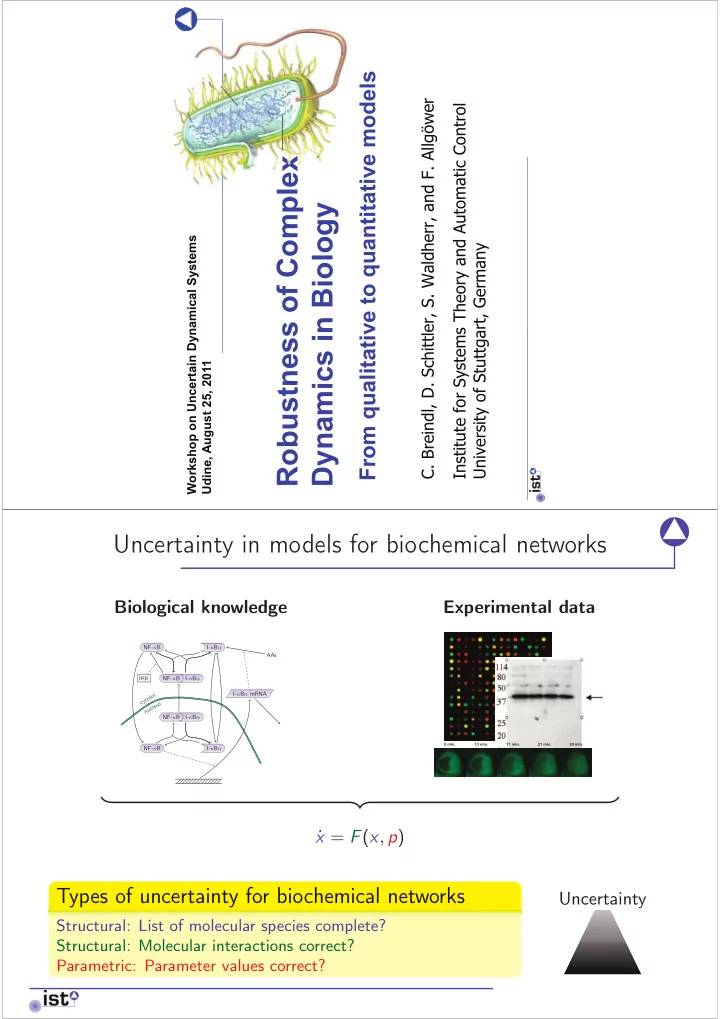
From qualitative to quantitative models C. Breindl, D. Schittler, S. Waldherr, and F. Allgöwer Institute for Systems Theory and Automatic Control Robustness of Complex Dynamics in Biology Workshop on Uncertain Dynamical Systems University of Stuttgart, Germany Udine, August 25, 2011 Uncertainty in models for biochemical networks Biological knowledge Experimental data NF- κ B I- κ B α AAs IKK NF- κ B I- κ B α I- κ B α mRNA cytosol nucleus NF- κ B I- κ B α NF- κ B I- κ B α x = F ( x , p ) ˙ Types of uncertainty for biochemical networks Uncertainty Structural: List of molecular species complete? Structural: Molecular interactions correct? Parametric: Parameter values correct?
Robustness on different levels x = F ( x , p ) ˙ Structural uncertainties Parametric uncertainties ¯ x A ? ? B C ? p Use assumption that nature favors How robust is the model’s behavior robust solutions to "identify" likely against perturbations in the interaction structure. parameters. F. Allgöwer, Robustness of complex dynamics in biology Outline Motivation 1 Steady state robustness of qualitative biological networks 2 Dynamical robustness of parametrized biological networks 3 Summary and conclusions 4 F. Allgöwer, Robustness of complex dynamics in biology
Outline Motivation 1 Steady state robustness of qualitative biological networks 2 Qualitative modeling framework Definition and computation of robustness measure Application example: Differentiation of mesenchymal stem cells Dynamical robustness of parametrized biological networks 3 Summary and conclusions 4 F. Allgöwer, Robustness of complex dynamics in biology Motivation Typical situation when studying gene regulatory networks: List of species (genes, mRNAs, ...) given. Only little is known about the interactions between these species. Mostly only qualitative measurements are available. Two important questions: 1 Can a given interaction structure explain the observed behavior in principle? 2 Given several alternative model structures: Which one is biologically more plausible? F. Allgöwer, Robustness of complex dynamics in biology
Important observed behavior: Multistability Some example systems Life or death decision Differentiation Pattern formation MSC fibroblast tripotent myoblast ... osteochondro- adipocyte progenitor pre-osteoblast chondrocyte Prakash and Wurst, osteoblast Cell.Mol.Life Sci. (2006) Multistability is a widely observed phenomenon. How to model and analyze the system when only qualitative information about the steady state behavior is available? F. Allgöwer, Robustness of complex dynamics in biology Modelling Framework: Network Structure System equations: ˙ x i = − k i · x i + ϕ i ( x ) , i ∈ { 1 , . . . , n } ϕ i ( x ) : arbitrary combination of sums and products of activation and inhibition functions Linear degradation A protein either activates or inhibits production of another protein ⇒ Monotonic activation- and inhibition functions Activation function Inhibition function act ( x 1 ) inh ( x 1 ) x 2 x 2 ⇒ ⇒ x 1 x 1 x 1 x 1 x 2 = − k 2 · x 2 + act ( x 1 ) ˙ x 2 = − k 2 · x 2 + inh ( x 1 ) ˙ Exact shapes of activation and inhibition functions not specified ⇒ structurally uncertain system F. Allgöwer, Robustness of complex dynamics in biology
Measurements x 2 x max 2 Steady state measurements X F 1 x high Variability between systems 2 Uncertain measurements ⇒ Representation of steady states as rectangular forward-invariant x low 2 sets F 2 x 1 ⇒ Only distinction between “high” x max 0 x low x high 1 1 1 and “low” Mutual inhibition network: ˙ x 1 = − k 1 · x 1 + inh 1 ( x 2 ) x 2 = − k 2 · x 2 + inh 2 ( x 1 ) ˙ Example for mutual inhibition network ] × [ x high F 1 = [ 0 , x low , x max ] = [ 0 , 0 . 2 ] × [ 0 . 8 , 1 ] 1 2 2 F 2 = [ x high , x max ] × [ 0 , x low ] = [ 0 . 8 , 1 ] × [ 0 , 0 . 2 ] 1 1 2 F. Allgöwer, Robustness of complex dynamics in biology Outline Question 1: Can a given interaction structure explain the observed steady state behavior in principle? F. Allgöwer, Robustness of complex dynamics in biology
Specifications Specification of (relative) concentrations and intervals ≤ x high 0 ≤ x low ≤ x max for all i i i i = [ x high I low = [ 0 , x low ] , I high , x max ] x i i x i i i Specification of m desired forward-invariant sets F z = I low / high × . . . × I low / high z = 1 , . . . , m x 1 x n Relates well to typical steady state measurements: Microarray data F. Allgöwer, Robustness of complex dynamics in biology Specifications Specification of (relative) concentrations and intervals ≤ x high 0 ≤ x low ≤ x max for all i i i i = [ x high I low = [ 0 , x low ] , I high , x max ] x i i x i i i Specification of m desired forward-invariant sets F z = I low / high × . . . × I low / high z = 1 , . . . , m x 1 x n Parametrization of activation/inhibition functions inh ( x ) N γ high - write ϕ � T if ϕ lies in the tube T - write ϕ � T if ϕ violates tube T γ low x low x high x max x 0 F. Allgöwer, Robustness of complex dynamics in biology
Conditions for forward-invariance Forward-invariance of an invariant set F (Nagumo) Consider the system ˙ x = g ( x ) . Assume that for each initial condition in a set X it admits a globally unique F solution. Let F ⊆ X be a closed and convex set . Then F is forward-invariant if and only if g ( x ) lies in the tangent cone to F in x for all x ∈ ∂ F . System satisfies assumptions. x 2 = − k 2 x low ˙ + inh ( x 1 ) 2 Every F z is hyper-rectangular. x 2 Functions ϕ are monotonic. x low 2 No nominal functions ϕ given F z x 1 x max x high 1 1 F. Allgöwer, Robustness of complex dynamics in biology Conditions for forward-invariance Forward-invariance of an invariant set F (Nagumo) Consider the system ˙ x = g ( x ) . Assume that for each initial condition in a set X it admits a globally unique F solution. Let F ⊆ X be a closed and convex set . Then F is forward-invariant if and only if g ( x ) lies in the tangent cone to F in x for all x ∈ ∂ F . System satisfies assumptions. x 2 = − k 2 x low ˙ + inh ( x 1 ) 2 Every F z is hyper-rectangular. x 2 Functions ϕ are monotonic. x low 2 ⇒ Only vertices have to be considered. No nominal functions ϕ given F z x 1 x max x high 1 1 F. Allgöwer, Robustness of complex dynamics in biology
Conditions for forward-invariance Forward-invariance of an invariant set F (Nagumo) Consider the system ˙ x = g ( x ) . Assume that for each initial condition in a set X it admits a globally unique F solution. Let F ⊆ X be a closed and convex set . Then F is forward-invariant if and only if g ( x ) lies in the tangent cone to F in x for all x ∈ ∂ F . System satisfies assumptions. x 2 = − k 2 x low ˙ + inh ( x 1 ) 2 Every F z is hyper-rectangular. x 2 Functions ϕ are monotonic. x low 2 ⇒ Only vertices have to be considered. γ low No nominal functions ϕ given F z ⇒ Reformulation using tubes as worst case approximation. x 1 x max x high 1 1 F. Allgöwer, Robustness of complex dynamics in biology Conditions for forward-invariance Invariance condition reformulated in terms of tubes x n with l i ∈ { low , high } and given tubes T i , k that satisfy Given a set F = I l 1 x 1 × . . . × I l n the conditions ∀ i ∈ { 1 , . . . n } : − k i · x i + γ i , 1 ◦ . . . ◦ γ i , q i ≥ 0 (1) where x i = min x i , and, with x j denoting the argument of ϕ i , k , x i ∈I li xi � l j 0 if ϕ i , k = ν i , k ∧ 0 ∈ I x j γ i , k = min { γ : ( x , γ ) ∈ T i , k ∧ x ∈ I l j x j } otherwise and ∀ i ∈ { 1 , . . . n } : − k i · x i + γ i , 1 ◦ . . . ◦ γ i , q i ≤ 0 (2) where x i = max x i , and, with x j denoting the argument of ϕ i , k , x i ∈I li xi γ i , k = max { γ : ( x , γ ) ∈ T i , k ∧ x j ∈ I l j x j } . If ∀ i , k : ϕ i , k � T i , k , then the set F is forward-invariant. F. Allgöwer, Robustness of complex dynamics in biology
The structure validation problem Consequence: Feasibility problem A given interaction structure can in principle explain the observed steady state behavior (forward-invariant sets) if and only if γ values (tubes) can be found such that Equations ( 1 ) and ( 2 ) hold. Often there are several model structures that can in principle explain the observed behavior. Can we find a measure for the biological plausibility of an interaction structure? Employ robustness considerations to develop this measure. F. Allgöwer, Robustness of complex dynamics in biology Outline Question 2: Find a measure for the biological plausibility of a model structure based on robustness considerations. F. Allgöwer, Robustness of complex dynamics in biology
Recommend
More recommend
Explore More Topics
Stay informed with curated content and fresh updates.