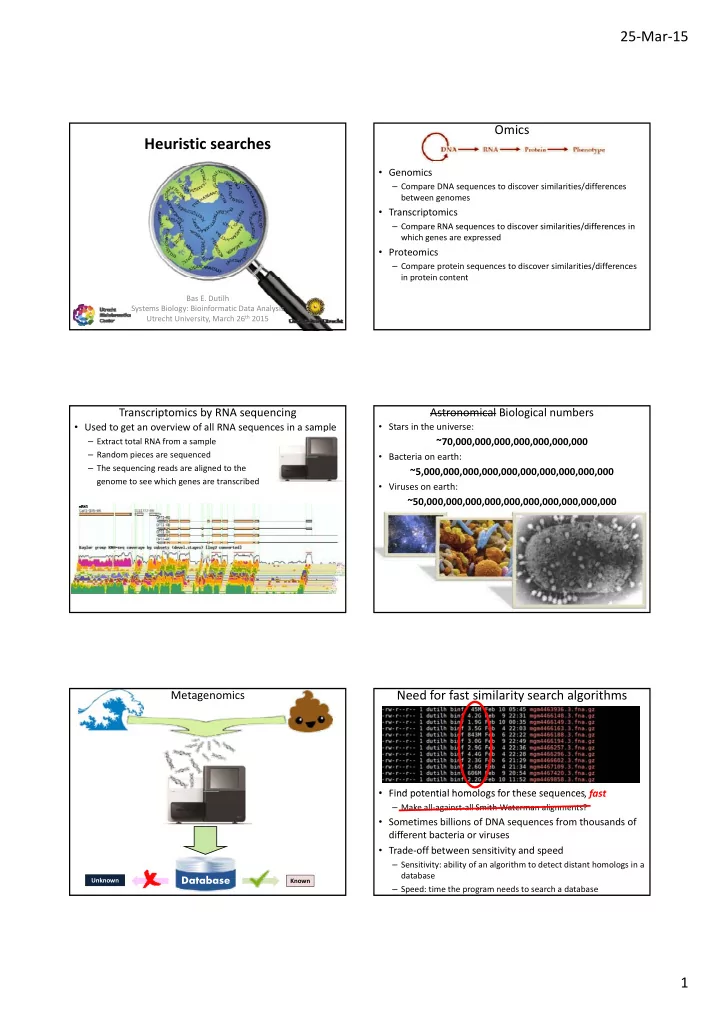

25 ‐ Mar ‐ 15 Omics Heuristic searches • Genomics – Compare DNA sequences to discover similarities/differences between genomes • Transcriptomics – Compare RNA sequences to discover similarities/differences in Compare RNA sequences to discover similarities/differences in which genes are expressed • Proteomics – Compare protein sequences to discover similarities/differences in protein content Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 26 th 2015 Transcriptomics by RNA sequencing Astronomical Biological numbers • Used to get an overview of all RNA sequences in a sample • Stars in the universe: – Extract total RNA from a sample ~70,000,000,000,000,000,000,000 – Random pieces are sequenced • Bacteria on earth: – The sequencing reads are aligned to the ~5,000,000,000,000,000,000,000,000,000,000 genome to see which genes are transcribed • Viruses on earth: ~50,000,000,000,000,000,000,000,000,000,000 Need for fast similarity search algorithms Metagenomics • Find potential homologs for these sequences • Find potential homologs for these sequences fast , fast – Make all ‐ against ‐ all Smith ‐ Waterman alignments? • Sometimes billions of DNA sequences from thousands of different bacteria or viruses • Trade ‐ off between sensitivity and speed – Sensitivity: ability of an algorithm to detect distant homologs in a database Database Unknown Known – Speed: time the program needs to search a database 1
25 ‐ Mar ‐ 15 k ‐ mer searches Degeneracy of the genetic code • Mutations in the 3 rd nucleotide of a codon often translate into the same amino acid (synonymous mutations) • k ‐ mers are “words” consisting of k nucleotides or amino acids – Discontiguous Megablast searches with spaced words containing – For k = 5, the amino acid sequence KAWSADV consists of the k ‐ mers: KAWSA, two out of every three nucleotides, allowing variations at the AWSAD, and WSADV third nucleotide of the codon • Identical words are easy to identify for a computer • Rule of thumb: – An index of the sequences can be stored in rapidly accessible memory (RAM) – Nucleotide sequences are the • However, this is not suitable to identify sequences at large , y q g least conserved evolutionary distance (many mutations) – Protein sequences are more conserved Sensitivity versus speed Basic Local Alignment Search Tool (BLAST) • Heuristic search algorithm – Makes shortcuts that are likely (but not guaranteed) to find the • (Almost) exact hits are easy to identify using fast k ‐ mer searches optimal hits – Not suitable for distant homologs • BLAST finds good potential homologs at reasonable speed – For example: which genes are expressed in a human cancer? • Here, the sequences can be matched to the human reference genome – 10 ‐ 50x faster than Smith ‐ Waterman – More than 100,000 queries per day on the NCBI BLAST server on the NCBI BLAST server • Terminology: – Query: sequence we search the database with • Highly diverged sequences (distant homologs) require careful, optimal alignment algorithms – Hit or Subject: similar sequence found in the database – This is slow: many algorithmic steps need to be performed by the central • BLAST is the most used bioinformatics program processing unit (CPU) of the computer – The BLAST article has been cited >54,000 times – For example: which unknown microbes are associated to coral disease? • Here, the sequences have to be compared with known microbial genomes (distantly related) BLAST input and output The BLAST search algorithm • Identifies potentially high ‐ scoring words ( k ‐ mers) in the query >p >pro rote tein in_s _seq eque uence_A BLAST input MTQSSHAVAA FD FDLGA LGAAL ALRQ RQE GL E GLTET TETDY DYSE SEI QR I QRDP DPNR NRAEL AELG TF TFGV GV (query sequences) – W = 3 for protein, W = 11 for DNA >pro >p rote tein in_s _seq eque uence_B MLTETDYSEI QR QRRLG RLGRD RDPN PNR AE R AELGM LGMFG FGVM VMN RA N RAEL ELGM GMFGY FGY – Based on substitution scores >pro >p rote tein in_s _seq eque uence_C MHAVAAFDLG AA AALRQ LRQEG EGLT LTE TD E TDYSE YSEIQ IQRR RRL GR L GRAM AMFG FGVMW VMWS EH EHCC CCYR YRNDD NDDA RPLL RP LLRP RPIK IKSP SP F FGAWVVIV • Quickly finds similar words in the database – All words in the database are indexed and stored in RAM, linked to similar “neighborhood words” • Extends seeds in both directions to find HSPs between query and hit – HSP: region that can be aligned with a score above a certain threshold BLAST output (hits) 2
25 ‐ Mar ‐ 15 BLAST flavors BLAST flavors: translated searches • Nucleotide ‐ nucleotide searches • We can exploit the higher – Nucleotide database, nucleotide query conservation of protein sequences – blastn (default: W = 11 nucleotides) when aligning DNA sequences, by • Find homologous genes in different species using translated searches – Megablast (default: W = 28 nucleotides) • Designed to efficiently find longer alignments between very similar nucleotide sequences • This allows for more sensitive searches that detect • Best tool to find highly identical hits for a query sequence homology at greater evolutionary distances • For example: find sequences from the same species – Discontiguous Megablast Discontiguous Megablast – For example: homologous genes in distantly related species – For example: homologous genes in distantly related species • Uses discontiguous words (e.g. W = 11 nucleotides: AT-GT-AC-CG-CG-T ) • blastx and tblastx first translate the nucleotide query into • For example, this can focus the search on codons (the third nucleotide of codons is less conserved due to the degeneracy of the gene � c code → next slide) protein before identifying high ‐ scoring words • Best tool to find nucleotide ‐ nucleotide hits at larger evolutionary distances for protein ‐ coding query sequences • tblastn and tblastx use a database of translated nucleotide • Protein ‐ protein searches sequences stored as proteins – Protein database, protein query sequences – blastp (default: W = 3 amino acids) • Find homologous proteins in different species The alignment bit ‐ score Expect value (E ‐ value) • For a given query, we are mostly interested in finding good • E ‐ value: how many times would you expect a hit this good, hits (highly similar, likely true homologs) by random chance • We could estimate this based on a score derived only from – Of course, this depends on the alignment score ( S ), the length of the query sequence ( m ), and the size of the database ( n ): the alignment like the bit ‐ score or percent identity S E Kmne – … but the chance of finding a hit with a high score by random chance increases if you use a larger database – K : constant for search space scaling – … so we have to correct for that … so we have to correct for that – λ : constant for substitution matrix correction λ f b i i i i Low E ‐ values are often given as exponents Low E ‐ values are often given as exponents • In the search below, we expect 10 ‐ 149 hits with a score of • In the search below, we expect 9.3 hits with a score of ≥ 436 by random chance ≥ 38.9 by random chance – Given the database size and query sequence length we expect – This is a lot, so this is a bad hit 0.00000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000001 hits by chance (this is not much, so this is a good hit) 3
Recommend
More recommend