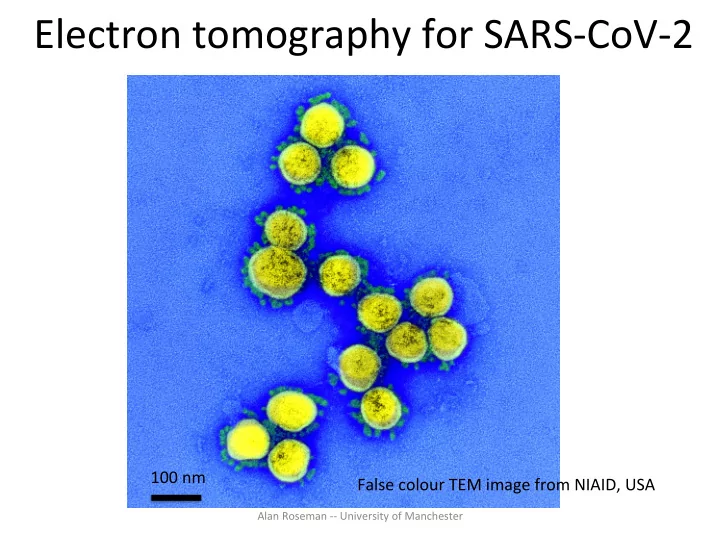

Electron tomography for SARS-CoV-2 100 nm False colour TEM image from NIAID, USA Alan Roseman -- University of Manchester
FEI Polara 300 kV TEM in FLS, UoM.
Nextstrain Alan Roseman -- University of Manchester
Alan Roseman -- University of Manchester
Nextstrain Alan Roseman -- University of Manchester
Nextstrain Alan Roseman -- University of Manchester
Nextstrain Alan Roseman -- University of Manchester
Alan Roseman -- University of Manchester
Model of virus and virus proteins Alan Roseman -- University of Manchester
TEM of SARS-CoV-2 (from NIAID) 100 nm Alan Roseman -- University of Manchester
TEM of SARS-CoV-2 TEM of SARS-CoV-2 (from NIAID) Alan Roseman -- University of Manchester
TEM of SARS-CoV-2 Alan Roseman -- University of Manchester TEM of SARS-CoV-2 (from NIAID)
Spike density at 20 Å resoluPon virus surface 25 nm Alan Roseman -- University of Manchester
Electron tomography for SARS-CoV-2 100 nm TEM of SARS-CoV-2 (from NIAID) Alan Roseman -- University of Manchester
Electron tomography
Sample preparaPon H. Saibil Alan Roseman -- University of Manchester
Cryo EM H. Saibil
EM ET principles • Parallel, coherent, electron beam • Missing wedge • Defocus/height dependent contrast transfer funcPon • Dose damage • DeformaPon/flow of sample • (correcPons for these are applied in current soVware, all would improve from local, spaPal, opPmisaPon) Alan Roseman -- University of Manchester
MHV cryoET, MHV is also a betacoronavirus 100 nm MHV (betacoronavirus) Alan Roseman -- University of Manchester
Issues with biological ET • Low SNR • Missing wedge • Progressive sample damage by beam • Unstable sample • High resoluPon obtained by averaging of idenPcal “moPfs” as sub-tomograms. Alan Roseman -- University of Manchester
Model virion with variable spikes Alan Roseman -- University of Manchester
Alan Roseman -- University of Manchester
Single parPcle workflow Alan Roseman -- University of Manchester
100 nm
Figure S3. Cryo-EM data processing workflow Cryo-EM structure of the 2019-nCoV spike in the prefusion Alan Roseman -- University of Manchester conformaCon . Wrapp et al, Science 2020.
Extended Data Figure 2 from: CryoEM image of isolated coronavirus spike proteins A.C. Walls, M.A. Tortorici, B.J. Bosch, B. Frenz, P.J.M. Ro_er, F. DiMaio, F.A. Rey, D. Veesler Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer Nature, 531 (2016), pp. 114-117 2D class averages Scale bars: 573 Å (micrograph) and 44 Å (class averages) Alan Roseman -- University of Manchester
WT cores WT fab labelled
360 Å 500 Å WT cores WT fab labelled
Back to those spikes …… Alan Roseman -- University of Manchester
ACE2 Adapted from: Vivian A. Scheuplein et al. J. Virol. 2015; doi:10.1128/JVI.03607-14 Alan Roseman -- University of Manchester
Spike density at 20 Å resoluPon The atomic models of the spike were determined by Walls et al (2020), using the cryoEM “single parPcle” method. Stabilised soluble spike proteins were produced in isolaPon, from mutated gene sequences, in lab grown cells. Structure, FuncCon, and AnCgenicity of the SARS-CoV-2 Spike Glycoprotein Alexandra C. Walls, Young-Jun Park, M. Alejandra Tortorici, Abigail Wall, Andrew T. McGuire, David Veesler Cell. 2020 Apr 16;181(2):281-292.e6. doi: 10.1016/j.cell.2020.02.058. Epub 2020 Mar 9. Alan Roseman -- University of Manchester
Spike density at 20 Å resoluPon Alan Roseman -- University of Manchester
Spike density at 20 Å resoluPon Alan Roseman -- University of Manchester
Spike density at 20 Å resoluPon Alan Roseman -- University of Manchester
Spike density at 20 Å resoluPon Alan Roseman -- University of Manchester
Spike density at 20 Å resoluPon Alan Roseman -- University of Manchester
Spike density at 20 Å resoluPon anPbody spikes virus surface Alan Roseman -- University of Manchester
ACE2 Alan Roseman -- University of Manchester
Model Main challenges: All general ET sample and dose issues menPoned. E.g. Missing wedge Defocus/height dependent contrast transfer funcPon Dose damage, low signal DeformaPon/flow of sample In addiHon: MulPple overlapping moPfs. Unknown level of variability. Alan Roseman -- University of Manchester
FEI Polara 300 kV TEM in FLS, UoM.
Viruses and the Development of QuanCtaCve Biological Electron Microscopy R.A. Crowther Notes Rec. R. Soc. Lond. 2004. Medical Research Council - Laboratory of Molecular Biology Cambridge Jo Butler, Samantha Wynne, John Berriman Helen Saibil
HIV trimer Nature, Vol 455|4 September 2008|doi:10.1038/nature07159
Test data and procedures to demonstrate steps and features to model for high resoluPon analysis This raw cryoEM data has been processed to resolve the capsid protein moPf to 3.1Å resoluPon, using sub-tomogram averaging. It has all the features of a good tomography dataset. The moPf is hexameric, and is probably not very heterogeneous. The challenge for coronaviurs is that in addiPon to a missing wedge, dose damage, etc; it has mulPple overlapping moPfs (the spikes) with an unknown level of variabiliy. Databank reference for images: EMPIAR-10164 hjps://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10164/ This HIV-1 capsid protein moPf has been resolved and analysed, and published 3 Pmes: Pub1. Schur, F. K. M. et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulaPng • assembly and maturaPon. Science 353, 506–508 (2016). 3.9Å, EMD-4015 Pub2. Turoňová, B., Schur, F. K. M., Wan, W. & Briggs, J. A. G. Efficient 3D-CTF correcPon for cryo- • electron tomography using NovaCTF improves subtomogram averaging resoluPon to 3.4Å. J. Struct. Biol. 199, 187–195 (2017). 3.4Å, EMD-3782 Pub3. Benjamin A. Himes & Peijun Zhang. emClarity: soVware for high-resoluPon cryo-electron • tomography and subtomogram averaging .Nature Methods, volume 15, pages. 955–961 (2018). 3.1Å, EMD-8986 Alan Roseman -- University of Manchester
Recommend
More recommend