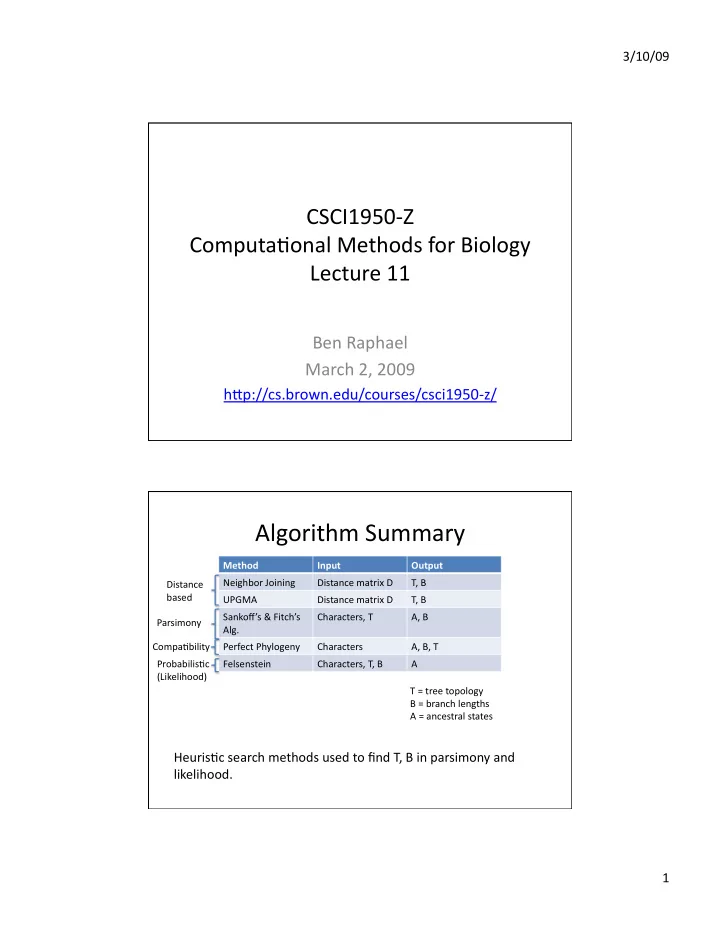

3/10/09 CSCI1950‐Z Computa3onal Methods for Biology Lecture 11 Ben Raphael March 2, 2009 hFp://cs.brown.edu/courses/csci1950‐z/ Algorithm Summary Method Input Output Neighbor Joining Distance matrix D T, B Distance based UPGMA Distance matrix D T, B Sankoff’s & Fitch’s Characters, T A, B Parsimony Alg. Compa3bility Perfect Phylogeny Characters A, B, T Probabilis3c Felsenstein Characters, T, B A (Likelihood) T = tree topology B = branch lengths A = ancestral states Heuris3c search methods used to find T, B in parsimony and likelihood. 1
3/10/09 Using Mul3ple Methods • Reliance on purely one method or dataset for phylogene3c analysis o_en provides incomplete picture. • If different methods (parsimony, distance‐ based, etc.) applied to same/different datasets give same result, greater confidence that this is correct answer. • Consensus or supertree methods can be used to combine this evidence. Phylogeny of Insects ( Nature 2003) Build phylogeny of winged and wingless s3ck insects Used data from: 18S ribosomal DNA (~1,900 base pairs (bp)) 28S rDNA (2,250 bp) Por3on of histone 3 (H3, 372 bp) Used mul3ple tree reconstruc3on techniques 2
3/10/09 Further Problems… Contradictory answers some3mes not a fault of data, but from overly simplis3c assump3ons about evolu3onary process. • No homoplasy: characters change state only once. • Independence of characters. • Modeling muta3ons in DNA. • Genes/genomes evolve only by single leFer muta3ons. Biology 101 3
3/10/09 Cell Division and Muta3on Single nucleo3de change Copy number Structural Whole‐Genome Phylogeny Finding same gene (descended from common ancestor) is non‐trivial. 4
3/10/09 Phylogeny of Insects ( Nature 2003) Build phylogeny of winged and wingless s3ck insects Used data from: 18S ribosomal DNA (~1,900 base These genes used because they are assumed pairs (bp)) to be highly conserved across large 28S rDNA (2,250 bp) evolu3onary distances. Por3on of histone 3 (H3, 372 bp) Used mul3ple tree reconstruc3on techniques Outline Whole Genome Phylogeny • Gene Trees vs. Species Trees • Reconciling Trees • Genome Rearrangements Genome sequencing is now rou3ne. Thus, data for these methods is increasingly available/ useful. 5
3/10/09 Gene Trees vs. Species Trees These trees indicate different phylogene3c rela3onships. One of them is wrong??? Gene Clusters/Families Gene duplica3on is a common mechanism for evolu3on of new gene func3on. (Ohno 1970) 6
3/10/09 Gene Trees and Species Trees Evolu3on of gene family inside species tree. Duplica3ons and losses occur. Gene Trees and Species Trees Hypothe3cal duplica3ons explain discrepancy between gene and species trees. 7
3/10/09 Gene Trees and Species Trees Duplica3ons are observed. Do not know which copies of gene descended from common ancestor. Evolu3on of Gene Tree Inside Species Tree Three events: 1. Specia3on 2. Loss 3. Duplica3on 8
3/10/09 Orthologs vs. Paralogs Three events: 1. Specia3on Orthologs : genes descended from a common ancestor. 2. Loss 3. Duplica3on Paralogs : genes related by duplica3on. Dis3nguishing orthologs from paralogs is difficult! Sequence similarity is not enough. Gene‐Species Tree Reconcilia3on Given : Rooted binary tree T G and rooted binary tree T S . Find : Embedding of T G in T S that minimizes number of duplica3ons (and losses). Embedded tree is called a reconciled tree (Goodman et al. 1979). 9
3/10/09 Reconcilia3on Example Reconcilia3on Example 10
3/10/09 Reconcilia3on Algorithm Zmasek and Eddy (2001) M(g) := λ G,T (g) Run Time analysis n = # leaves in T G Ini3aliza3on: O( n ): number nodes of T S O( n ): label external nodes (using hash‐table) Reconcilia3on Algorithm O( n ) O( n log n ) O( n 2 ) worst case. O( n 2 ) Using algorithms to compute LCA in O(1) 3me gives O( n ) algorithm (Zhang 1997, Chen et. al 2001) 11
3/10/09 Gene Trees and Species Trees Extensions 1. Species tree T S unknown. Use minimum duplica3on/loss as objec3ve func3on – to search tree space. – NP‐hard (Ma et al. 1998) – Heuris3c search (NNI, SPR, TBR, etc.) 2. Mul3ple gene trees T G 1 , T G 2 , …, T GN N � c ( T G i , S ) Minimize: i =1 Where c( T Gi , S) = # duplic./losses on reconciled tree for T Gi . 12
3/10/09 Roo3ng By Duplica3on • Gene trees o_en unrooted. • Root determined using outgroup: species known to be distantly related to all remaining. • Duplica3ons can be used to determine outgroup. 1 duplica3on 3 duplica3ons Roo3ng By Duplica3on Tree of life: Three major branches: bacteria, archaea, eukaryotes. No outgroup! 13
Recommend
More recommend