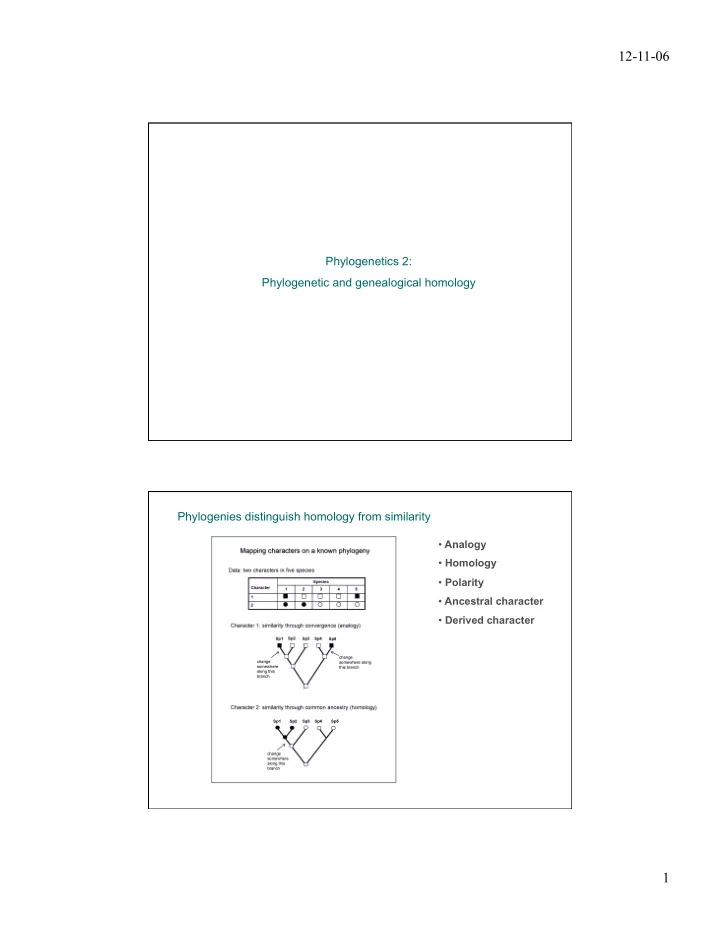

12-11-06 Phylogenetics 2: Phylogenetic and genealogical homology Phylogenies distinguish homology from similarity • Analogy • Homology • Polarity • Ancestral character • Derived character 1
12-11-06 Alignment of mammalian beta-globin gene sequences human cow rabbit rat opossum GTG CTG TCT CCT GCC GAC AAG ACC AAC GTC AAG GCC GCC TGG GGC AAG GTT GGC GCG CAC ... ... ... G.C ... ... ... T.. ..T ... ... ... ... ... ... ... ... ... .GC A.. ... ... ... ..C ..T ... ... ... ... A.. ... A.T ... ... .AA ... A.C ... AGC ... ... ..C ... G.A .AT ... ..A ... ... A.. ... AA. TG. ... ..G ... A.. ..T .GC ..T ... ..C ..G GA. ..T ... ... ..T C.. ..G ..A ... AT. ... ..T ... ..G ..A .GC ... GCT GGC GAG TAT GGT GCG GAG GCC CTG GAG AGG ATG TTC CTG TCC TTC CCC ACC ACC AAG ... ..A .CT ... ..C ..A ... ..T ... ... ... ... ... ... AG. ... ... ... ... ... .G. ... ... ... ..C ..C ... ... G.. ... ... ... ... T.. GG. ... ... ... ... ... .G. ..T ..A ... ..C .A. ... ... ..A C.. ... ... ... GCT G.. ... ... ... ... ... ..C ..T .CC ..C .CA ..T ..A ..T ..T .CC ..A .CC ... ..C ... ... ... ..T ... ..A ACC TAC TTC CCG CAC TTC GAC CTG AGC CAC GGC TCT GCC CAG GTT AAG GGC CAC GGC AAG ... ... ... ..C ... ... ... ... ... ... ... ..G ... ... ..C ... ... ... ... G.. ... ... ... ..C ... ... ... T.C .C. ... ... ... .AG ... A.C ..A .C. ... ... ... ... ... ... T.T ... A.T ..T G.A ... .C. ... ... ... ... ..C ... .CT ... ... ... ..T ... ... ..C ... ... ... ... TC. .C. ... ..C ... ... A.C C.. ..T ..T ..T ... Homology: similarity among two or more individuals or lineages in a feature/character, or character-state, that is the result of inheritance from a common ancestor Homologous character verses Homologous character state ACG TAC TAA ACG TAC TAA C C ACG TAT TAA ACG TAT TAA DNA alignment T T ACG TAT TAA ACG TAT TAA Molecular evolution: positional homology 2
12-11-06 Phylogenies distinguish homology from similarity Phylogenetic perspective on homologous characters and homologous character states ACG TAC TAA S YNAPOMORPHY : a shared derived character state in C two or more lineages. ACG TAT TAA These must be homologous in state. T This represents a true phylogenetic similarity ACG TAT TAA Ancestral character states C ⇒ T ACG TAT TAA APOMORPHY : a derived character state unique to a single lineages C ACG TAC TAA C ACG TAC TAA Ancestral character states Implicit in the above alignment is the assumption of positional homology for the “red” position above. At this position C and T are non-homologous character states. Note that the pair of T’s in the first example , and the pair of C’s in the second example represent homologous character states. Phylogenies distinguish homology from similarity It is possible for character-states to be identical but non- homologous (Homoplasy) 1. Convergence 2. Reversal 3. Parallelism Homoplsy arises in molecular datasets when multiple substitutions occur at a single site. - “ multiple substitutions ” - “ multiple hits ” - “ superimposed substitutions ” 3
12-11-06 Phylogenies distinguish homology from similarity: convergence Convergent (non-phylogenetic) similarity of nucleotide character states; i.e., homoplasy T ⇒ C ⇒ A ACG GAA TAA ACG GAT TAA T T ACG GAG TAA G G ⇒ A ACG GAA TAA Phylogenies distinguish homology from similarity: reversal Reversal leads to non-homologous similarities in character state. A ⇒ C ⇒ A ACG GAA TAA ACG GAA TAA A A ACG GAT TAA T ACG GAT TAA 4
12-11-06 Phylogenies distinguish homology from similarity: parallelism Parallel evolution leads to non-homologous similarities in character states. A ⇒ T ACG GAT TAA ACG GAA TAA A A ⇒ T A ACG GAT TAA A ACG GAA TAA Evolutionary dissociation “ A change in linkage or effects that particular traits from different levels of biological organization have on each other over evolutionary time ” ⎯ Meyer and Mindell (2001) 5
12-11-06 Evolutionary dissociation The phylogenetic concept of homology allows distinction between homologous characters and non-homologous characters states in the case of evolutionary dissociation. Dissociation events G ENE A is homologous as a molecular character in all five lineages G ENE A’s functional role in development also is homologous in all five lineages The “character state” of G ENE A is NOT homologous in lineage 5 as compared with lineages 1 and 2 even though they have the same role! This is another example of homologous characters with non-homologous character states. Adapted from: Mindell and Meyer (2001) Trends Ecol Evol. (16) 434-440. Role 2: morphologically similar structures with different molecular evolutionary basis Evolutionary dissociation Butterfly embryo Sea urchin larva Homologous genes control the basis for non-homologous structures. Apparently, Genetic co-option has occurred. Gene duplication provides one mechanism; non- homologous character states within homologous positions of a gene provides another. 6
12-11-06 Alternatives to phylogenetic concept of homology Serial Homology refers to repeated structural units of morphology of an organism. • It seems reasonable that such units are derived from homologous genes or developmental processes; whether this assumption turns out to be generally applicable to morphological features exhibiting serial homology has not yet been demonstrated. Functional homology is generally used when the functional attributes of organisms are similar due to shared ancestry. • However, due to a long history of using this term to denote simple similarity, one should not assume this usage unless unequivocally stated. Biological homology is defined by Wagner (1989) as morphological structures that share “developmental constraints, caused by locally acting self-regulating mechanisms or organ differentiation”. • Although phylogeny can be used to help diagnose such homology, the definition does not explicitly include the condition of shared ancestry. Alternatives to phylogenetic concept of homology percent homology For DNA (or amino acid) sequences use percent similarity 7
12-11-06 Trees within trees Trees within trees: reticulation Conventional representation Conventional representation Path 1 Path 1 Path 2 Path 2 A A B B A A A B B B C C D D C C C C C C D D D D D D E E F F E E E F F F E E E F F F I I I I I I I I 8
Species 1 Species 1 Trees within trees Trees within trees Species 2 Species 2 Species 3 Species 3 Species 4 Species 4 12-11-06 9
Species 1 Species 1 Trees within trees Trees within trees Species 2 Species 2 Species 3 Species 3 Species 4 Species 4 12-11-06 10
12-11-06 Trees within trees Species 1 Species 3 Species 2 Species 4 Population genetics: coalescent theory to study populations Trees within trees Drift Selection time Population history 11
12-11-06 Trees within trees Polymorphism and substitution (highly simplified) along a branch of a phylogeny Residence time: the time that Mean residence time is a particular neutral determined by effective polymorphism is present in a population size (N e ) Coalescent population. Time Population substitution 1 Population substitution 2 Population substitution 3 A ⇒ C A ⇒ G C ⇒ A A ⇒ C ⇒ A ⇒ G GAG GAC A A GAT T GAT Forms of homology (for genes) 1 . O RTHOLOGY . Orthologous genes are derived from the divergence of an organismal lineage; i.e., a speciation event. Thus if we look at orthologs on a phylogeny we see that their most recent common ancestor represents the coalescence of two organismal lineages. 2. P ARALOGY . Paralogous genes are derived from the divergence event within a genomes; i.e., a gene duplication event. In this case if we look at paralogs on a phylogeny we see that they coalesce at a gene duplication event. 3. P RO - ORTHOLOGY . A gene is pro-orthologous to another gene if they coalesce at a speciation event that predates a gene duplication event. Thus a single-copy gene in organism A is pro- orthologous to a gene that is present in multiple copies in organism B due to gene duplication events that followed the divergence of organisms A and B. 4. S EMI - ORTHOLGY . This is a term that simply takes the reverse perspective of pro-orthology. Any one of the multi-copy genes in the genome of organism B is said to be semi-orthologous to a single copy gene in the genome of organism A, if the most recent common ancestors of those genes coalesce at a point in time that predates the gene duplication event. 5. P ARTIAL HOMOLOGY . This refers to the situation that arises when the evolutionary histories of different segments within the same gene coalesce at different ancestors. This can arise from evolutionary processes such as homologous recombination or exon shuffling. 6. G AMETOLOGY . Gametologs coalesce at an event that isolated those genes on opposite sex chromosomes; i.e., they coalesce at the point when they became isolated from the process of recombination. 7. X ENOLOGY Genes that coalesce at either a speciation or duplication event, but whose evolutionary histories do not fit with that of the organismal lineages which carry such genes due to one or more lateral gene transfer events. 8. S INOLOGY . Homologous genes found within the same organism’s genome have different evolutionary histories due to the fusion of formerly evolutionarily independent genomes, such as in endosymbiosis. 12
Recommend
More recommend