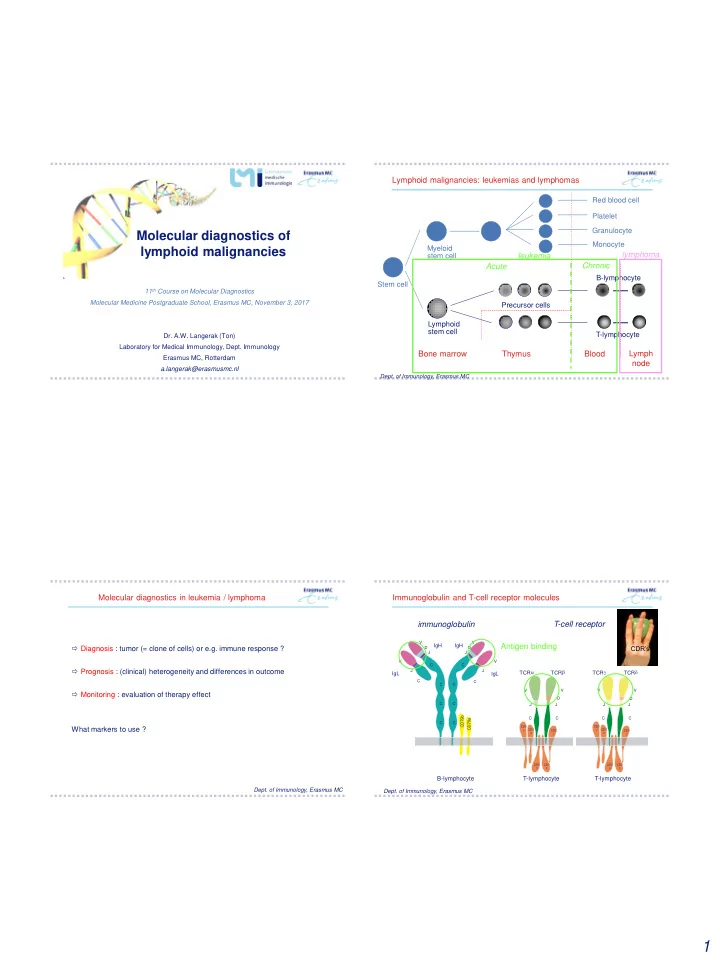

Lymphoid malignancies: leukemias and lymphomas Red blood cell Platelet Granulocyte Molecular diagnostics of Monocyte lymphoid malignancies Myeloid lymphoma stem cell leukemia Chronic Acute B-lymphocyte Stem cell 11 th Course on Molecular Diagnostics Molecular Medicine Postgraduate School, Erasmus MC, November 3, 2017 Precursor cells Lymphoid stem cell T-lymphocyte Dr. A.W. Langerak (Ton) Laboratory for Medical Immunology, Dept. Immunology Bone marrow Thymus Blood Lymph Erasmus MC, Rotterdam node a.langerak@erasmusmc.nl Dept. of Immunology, Erasmus MC Molecular diagnostics in leukemia / lymphoma Immunoglobulin and T-cell receptor molecules immunoglobulin T-cell receptor V V IgH IgH Antigen binding Diagnosis : tumor (= clone of cells) or e.g. immune response ? CDR’s D D J J V V C C Prognosis : (clinical) heterogeneity and differences in outcome J J a b g d TCR TCR TCR TCR IgL IgL C C C C V V V V Monitoring : evaluation of therapy effect D D C C J J J J b 9 a C C C C 7 9 C C D 7 C D What markers to use ? C CD3 CD3 e CD3 CD3 e CD3 CD3 d g d g CD3 CD3 CD3 CD3 x x x x B-lymphocyte T-lymphocyte T-lymphocyte Dept. of Immunology, Erasmus MC Dept. of Immunology, Erasmus MC 1
IG / TCR heterogeneity : combinatorial and junctional diversity IG / TCR heterogeneity : junctional diversity m V D J s C H H H 1 2 3 4 5 6 66 1 2 3 4 27 1 2 3 4 5 6 IGHV3-21 IGHD3-3 IGHJ4-1 IGHCM D J joining junctional region V D-J joining IGHV3-21 (germline) insertion IGHD3-3 (germline) insertion IGHJ4-1 (germline) V V IgH IgH D D J J TGTATTACTGTGCGAGA GTATTACGATTTTTGGAGTGGTTATTATACC ACTACTTTGACTACT V V C C TGTATTACTGT CGATTTTTGGAGTGGTTATTATA AGGC GTCCA TGACTACT J J transcription IgL IgL TGTATTACTGTGCG TTA TATCCGGA CGATTTTTGGAGTGGTTATTATAC CGATCG CTTTGACTACT C C TGTATTACTGTGC TTTTGGAGTGGTTATTATACC ACTACTTTGACTACT C C CCGGACTG GGT precursor IGH mRNA TGTATTACTGTGCGAG TATTACGATTTTTGGAGTGGTTAT TTTGACTACT CTGAGTC CGTAGCGTA TGTATTACTG CGATTTTTGGAGTGGTTATTATA ACTTTGACTACT ACATCGA CGTAG C C TGTATTACTGTGCG TACGATTTTTGGAGTGGTTATTAT TGACTACT CGT GGCTAAGG RNA splicing b translation TGTATTACTGTGCGA TTACGATTTTTGGAGTGGTTATTATACC CCGG CGGAGC TACTTTGACTACT 9 a 7 9 m C C D 7 V D J C D TGTATTACT GATG GATTTTTGGAGTGGT GGTTC ACTACTTTGACTACT C C mature IGH mRNA TGTATTACTGTGCG ATTACGATTTTTGGAGTGGTTATTATA TTCA CGATCGA CTTTGACTACT TGTATTACTGTGC TTTGGAGTGGTTATTATA CC ACTTTGACTACT TGTATTACTGTGCGAGA ATTACGATTTTTGGAGTGGTTATTATACC GACTACT GGCTAG GTCG TGTATTACTGTG TATTACG ATTTTTGGAGTG CTACTTTGACTACT GTCCAG CCGTAG TGTATTA CGATTTTTGGAGTGGTTATTATA ACT CCGGA C TGTATTACTGTGC ACGC ATTACGATTTTTGGAGTGG GTACG TTTGACTACT TGTATTACTGTGCGA GTATTACGATTTTTGGAGTGGTTATTATACC CGTA GGCA ACTTTGACTACT Dept. of Immunology, Erasmus MC Dept. of Immunology, Erasmus MC Estimated potential primary repertoire of human IG / TCR molecules IG / TCR rearrangements in leukemia and lymphoma TCR ab TCR gd Ig Ig TCR a TCR b TCR g TCR d IgH Ig genes rearranged Number of genes - V genes > 100 > 50 > 40 > 50 > 70 6 6 B-lymphocyte - D genes 27 - - - 2 - 3 - J genes 6 5 4 55 13 5 4 Precursor cells Combinatorial Lymphoid > 5 x 10 6 > 5 x 10 6 > 5000 diversity stem cell T-lymphocyte TCR genes rearranged Junctional region ++ + ++ ++ ++++ diversity Lymph Bone marrow Thymus Blood node Estimation of total normal B/T cells: unique IG / TCR gene rearrangement primary receptor > 10 12 > 10 12 > 10 12 repertoire lymphoid tumor: all cells will have identical Ig or TCR genes (monoclonal) Dept. of Immunology, Erasmus MC Dept. of Immunology, Erasmus MC 2
Molecular diagnostics in leukemia / lymphoma PCR-based analysis of Ig / TCR rearrangements m V H D H J H s C 1 2 3 4 5 6 66 1 2 3 4 27 1 2 3 4 5 6 Diagnosis : tumor (= clone of cells) or e.g. immune response ? DH JH joining Clonality testing using IG / TCR rearrangements, as polymorphic markers VH DH-JH joining PCR-based fragment analysis / spectratyping • non-hemopoietic tissue : Ig / TCR segments far apart no PCR products • normal BM, PB, lymph node : Ig / TCR segments coupled PCR products further discrimination between polyclonality and monoclonality: GeneScan fragment analysis : length of PCR products Dept. of Immunology, Erasmus MC Dept. of Immunology, Erasmus MC Clonality testing CDR3 heterogeneity Clonality testing CDR3 heterogeneity fluorescence fluorescence intensity intensity dominant peak = clone DNA fragment length DNA fragment length Dept. of Immunology, Erasmus MC Dept. of Immunology, Erasmus MC 3
IGH gene analysis : multiplexing IGH gene analysis : multiplexing 27 6 >100 7 IGHV families Leukemia 2003;17:2257-2317 Somatic hypermutation can hamper primer annealing Complementarity of IG targets secondary B-cell lymphoma antibodies bind antigen clearance responses DLBCL (n=116) FCL (n=109) plasma cells - FR1 67 % - FR1 73 % memory SHM: B-cells V H3-21 D H3-3 J H4-1 - FR2 57 % - FR2 76 % FR1 CDR1 FR2 CDR2 FR3 CDR3 T IGH class T switch FR1 CDR1 FR2 recombination T T - FR3 47 % - FR3 52 % TCCTGTGCAGCCTCT GGATTCACCTTCAGTAGCTATAGC ATGAACTGGGTCCGCCAGGCTCCAGGGAAG T (CSR) T TCCTGTG AGCCTCT T GGATTCACCTTCAGTAGCTATAGC ATGAACTGGGTCCGCCAGGCTCCAGGGAAG TCCTGTGCAGCCTCT GGATTCACCTTCAGTAGCTATAGC ATGAACTGGGTCCGCCAGGCTCCAGGGAAG positive selection TCCTGTGCAGCCTCT GGATTCACCTTCAGTAGCTATAGC ATGAACTGGGTCCGCCAGGCTCCAGGG AG G - all FR 77 % - all FR 84 % of B-cells for TCCTGTGCAGCCTCT GGATTCAC TTCAGTAGCTATAGC G ATGAACTGGGTCCGCCAGGC CCAGGGAAG A DC ATGAACTGGGTCCGCCAGGCTCCAGGGAAG DC TCCTGTGCAGCCTCT GGATTCACCTTCAGTAGCTATAGC antigen binding TCCTGTGCAGCCTCT GGATTCACCTTCAGTAG TATAGC T ATGAACTGGGTCCGCC GGCTCCAGGGAAG G DC TCCTGTGCAGCCTCT GGATTCACCTTCAGTAGCTATA C A ATGAA TGGGTCCGCCAGGCTCCAGGGAAG A TCCTGTGCAGCCTCT GGATTCACCTTCAGTAGCTATAGC ATGAACTGGGTCCGCCAGGCTCCAGGGAAG T T selection for B-lymphocytes with high-affinity antibodies T naive B-cell proliferations TCCTGTGCAGCCTCT GGATTCACCTTCAGTAGCTAT GC G ATGAACTGGGTCCGCCAGGCTCCAGGGAAG T ATGAACTGGGTCCGCCAGGCTCCAGGGAAG (virgin) TCCTGTGCAGCCTCT GGATTCACCTTCAGTAG TA ATA CT and somatic T B-cells hypermutation (SHM) DLBCL: Diffuse large B-cell lymphoma; FCL: Follicular Cell Lymphoma Lymph node follicle Physiology: antibody affinity maturation Diagnostics: primer misannealing Dept. of Immunology, Erasmus MC Leukemia 2007;21:207-214 4
Molecular diagnostics in leukemia / lymphoma Complementarity of IG targets B-cell lymphoma DLBCL (n=116) FCL (n=109) Prognosis : (clinical) heterogeneity and differences in outcome - FR1 67 % - FR1 73 % Somatic mutation analysis of rearranged IG genes - FR2 57 % - FR2 76 % - FR3 47 % - FR3 52 % Sequencing - all FR 77 % - all FR 84 % - all FR + D H -J H 83 % - all FR + D H -J H 86 % - IGH + IGK 91 % - IGH + IGK 100 % DLBCL: Diffuse large B-cell lymphoma; FCL: Follicular Cell Lymphoma Alternative targets: - chromosome aberrations / translocations / deletions mutations / SNV’s - Clonality testing possible in vast majority of patients Leukemia 2007;21:207-214 Dept. of Immunology, Erasmus MC B-cell chronic lymphocytic leukemia (CLL) IGHV mutational analysis • IGH FR1 BIOMED-2 multiplex PCR + sequencing • Tumor of mature circulating B-cells • blast in IMGT database • determine V, D, J gene usage, reading frame • High leukocyte counts • Monomorphic small round lymphocytes Clinical features • Most patients asymptomatic • Others show fatigue, splenomegaly, lymphadenopathy • Clinical course mostly indolent, but can also be more aggressive search for prognostic factors !! ~1995 : somatic hypermutation (SHM) status of IGHV gene: unmutated: poor prognosis mutated: favorable prognosis Dept. of Immunology, Erasmus MC 5
IGHV mutational analysis Survival according to IGHV mutational status • determine total number of mutations Erasmus MC CLL cohort • calculate % homology : # mutations / # bases in V gene Overall survival mutation (%germ < 98) 100 yes 98-100% homology Cumulative percentage to closest IGHV 75 = non-mutated 50 no 25 N F no 52 17 y es 82 6 Logrank P<.001 <98% homology 0 to closest IGHV 60 0 12 24 36 48 months 60 At risk: = mutated 0 no 52 26 16 7 3 0 0 y es 82 54 27 10 6 0 Haematologica 2006;91:56-63 Stereotypy of Ig receptors and clinical impact Molecular diagnostics in leukemia / lymphoma Monitoring : evaluation of therapy effect Minimal residual disease analysis using patient-specific IG/TCR rearrangements real-time quantitative PCR (RQ-PCR) IG homology in various CLL patients: Ghia et al, Blood 2005 Antigen-driven pathogenesis? Tobin et al., Blood 2002, 2003 Dept. of Immunology, Erasmus MC 6
Recommend
More recommend